
Explore our Protein Kinase solutions
Biochemical kinases
A universal solution: KinEASE assays
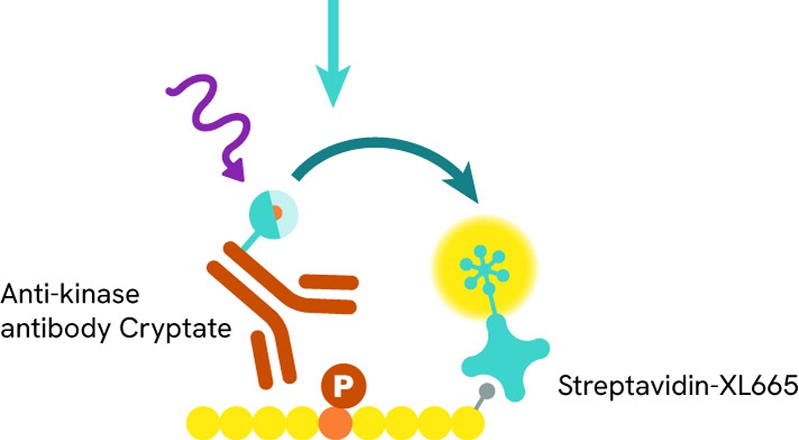
Our experts developed specific HTRF™ KinEASE kits to investigate kinase activity, characterize kinases, and screen for inhibitors. The five kits in the product range offer a semi-universal method for addressing phosphorylation on Serine/Threonine and Tyrosine residues, respectively. Over 272 kinases have already been validated with this method.
The principle for these assays is divided into two main steps:
Enzymatic step: During the enzymatic step, the tagged substrate is incubated with the kinase of interest and ATP, allowing the phosphorylation reaction to begin.
Detection step: Detection reagents are added and recognize the phosphorylated substrate. The resulting Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal is proportional to the phosphorylation level.
Suggested content
- Measuring kinase activity under nearly any condition
- Addressing multiple therapeutic areas with HTRF KinEASE assays
A substrate-specific solution: LANCE™ Ultra kinase assay
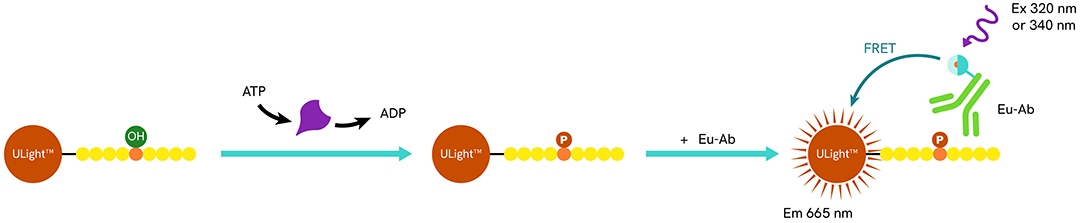
LANCE technology (LANthanide Chelate Excitation) Ultra is a solution in which your fluorescently labeled substrate can be chosen specifically. Over 300 kinases were tested and validated with this approach.
The assay protocol is divided in two steps:
- Enzymatic step: The ULight™-peptide substrate is phosphorylated in the presence of kinase and ATP.
- Detection step: The substrate is captured by an Eu-anti-phospho-substrate antibody. Upon excitation, the Eu-chelate transfers its energy to the ULight dye, resulting in fluorescent light emission at 665 nm.
Kinase binding platform
Why choose the HTRF kinase binding assay?
One of the most well-known kinase inhibitors, Gleevec (Imatinib), binds to a non-activated form of Abl kinase. Inhibitors like Gleevec are challenging to detect using activity-based assays. Therefore, setting up kinase binding assays for your kinase of interest is advantageous to ensure you don’t miss out on detecting such compounds.
HTRF kinase binding assays are simple to set up, utilizing a convenient mix-and-measure detection format. These assays do not require the presence of ATP. Instead, they measure the direct displacement of the fluorescent inhibitor from the ATP binding pocket. This allows you to determine the affinity of your inhibitor by performing dose-response curves.
How it works: assay principle
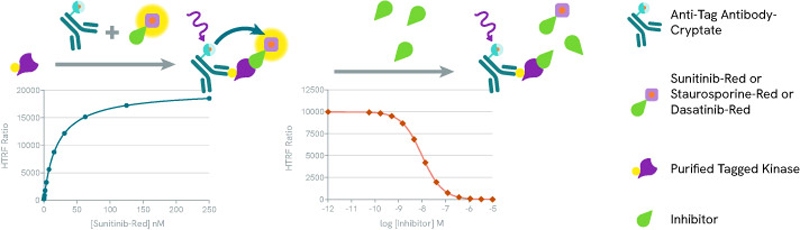
The HTRF Kinase binding assay is based on a biochemical sandwich format. When a tagged kinase is present, an HTRF signal is generated when the tracer is bound to the kinase. Upon adding competitive inhibitors, the fluorescent Staurosporine (or Dasatinib or Sunitinib) is displaced and the HTRF signal disappears.
Kinase inhibitor selection for kinase binding assays
The Revvity discovery kit includes three fluorescent ATP-competitive kinase inhibitors: Staurosporine-red, Dasatinib-red, and Sunitinib-red. We estimate that these three fluorescent inhibitors cover at least 80% of the human kinome.
Our scientists have written three application notes, one per tracer, to guide you in your studies. Each application note provides validation data, guidance for Kd and Ki determination, and case studies.
- HTRF Kinase Binding Assays: Sunitinib-Red Validation
- HTRF Kinase Binding Assays: Dasatinib-Red Validation
- HTRF Kinase Binding Assays: Staurosporine-Red Validation
Kinetic binding parameters: determining Kon and Koff
Dissociation kinetics, and thus inhibitor residence time, have become key parameters for predicting in-vivo drug efficacy outcome. HTRF kinase binding platform assays are ideal for studying the binding kinetics of inhibitors.
Get complete information in this application note entitled, “Kinetic binding of kinase inhibitors and determination of Kon, Koff rates”.
Suggested content
Featured resources
































