Extraction to analysis
Metagenomics workflows: 16 sequencing, WGS, and metatranscriptomics

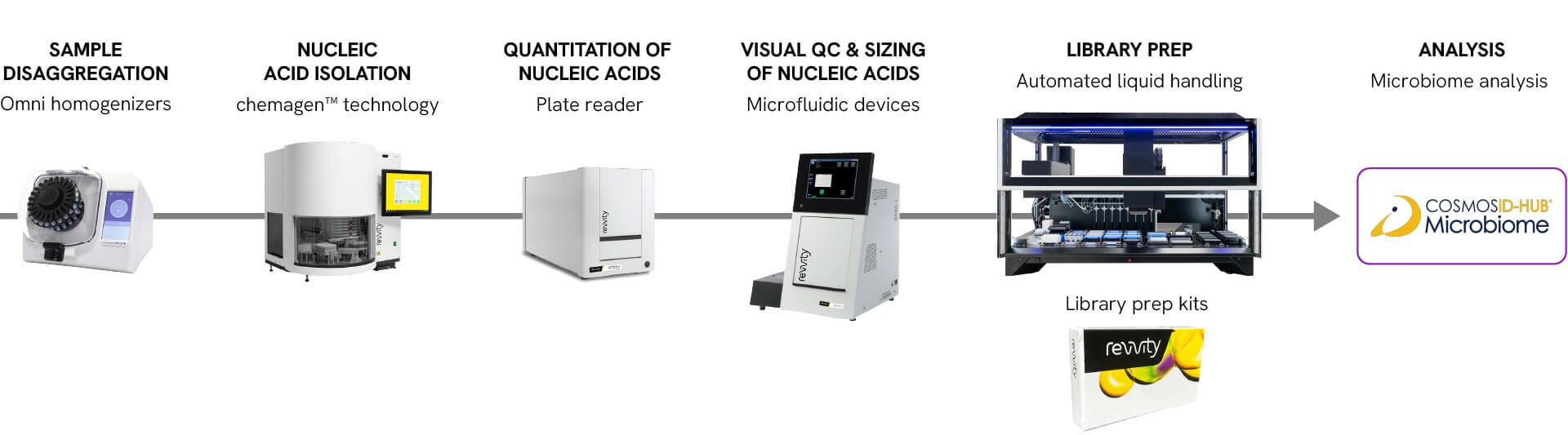
Sample disaggregation
Omni homogenizers

Omni Bead Ruptor bead mills, built on over 60 years of sample prep expertise, are indispensable powerhouse units for labs looking to transition from single, manual handheld preps to multi-sample processing for metagenomic workflows.
Nucleic acid isolation
chemagen™ technology

Nucleic acid extraction is a critical precursor to library preparation. High-quality, pure nucleic acids are essential for robust libraries. Discover the chemagic™ kits used on the chemagic instruments enabling a reliable nucleic extraction workflow for your metagenomic analysis.
Quantitation of nucleic acids
Plate reader

Quantitation of DNA and RNA prior to sequencing is an important step in the NGS library preparation workflow. Using the VICTOR Nivo™ plate reader to quantify samples and final libraries significantly reduces measurement time and increases throughput.
Visual QC sizing of nucleic acids
Microfluidic devices

Improve your nucleic acid analysis with the LabChip® automated microfluidic capillary electrophoresis (micro-CE) technology and enable the analysis of genomic DNA and NGS libraries in seconds. Reporting is improved and simplified through the automatic generation of data as an electropherogram, virtual gel, or tabular report.
Automated library prep
Automated liquid handling

Revvity NGS liquid handlers' standardized deck configuration allows for complete liquid handling, thermal control, and waste management for the most complex application requirements in next generation sequencing sample preparation protocols. Revvity's liquid handler portfolio has validated protocols for several metagenomics NGS applications, including shotgun and 16S sequencing.
Reagents
Library prep kits

Revvity offer library prep solutions for your metagenomics needs including 16S rRNA sequencing, shotgun sequencing, and metatranscriptomics solutions enabling the analysis of microbial communities without culturing.
16S rRNA sequencing is a cost-effective NGS tool for identifying and comparing bacterial species in a sample. It uses the highly conserved 16S rRNA gene for genus and species identification, making it ideal for microbial profiling and phylogenetic studies.
Shotgun metagenomic sequencing samples all genes in a complex sample, assessing bacterial diversity and identifying microbial abundance and functional analysis.
Revvity also provides metatranscriptomics solutions, including directional RNA-seq library prep and CRISPR-Cas9 ribodepletion kits, to enhance the detection of biologically relevant transcripts in complex communities.
CosmosID-HUB
Online microbiome software

Select NEXTFLEX kits are available bundled with access to CosmosID-HUB, an online software solution that enables fast and easy analysis of complex microbiome data.
The CosmosID-HUB gives scientists user-friendly access to version-controlled and validated 16S and shotgun sequencing pipelines.
The machine-learning powered software also enables rapid data interpretation through a comparative analysis software.
Meet the experts.
Join Britton Strickland, Ph.D., and Gustav Lindved in the webinar, Advanced, End-To-End Shotgun Metagenomics Workflows, as they introduce the CosmosID-HUB and how it enables the understanding of the functional roles of microbes in various environments.
Sample homogenization
Jumpstart your infectious disease research with a robust and powerful homogenizer designed to simplify sample preparation and prepare high quality lysates prior to nucleic acid extraction and metagenomic workflows.
Suggested links:
Isolation of nucleic acids from complex samples
Nucleic acid extraction is a critical precursor to library preparation. High-quality, pure nucleic acids are essential for robust libraries. Discover the chemagic™ kits used on the chemagic instruments enabling a reliable nucleic extraction workflow for your metagenomic analysis.
Suggested links:
Genomic DNA and NGS library analysis
Improve your nucleic acid analysis with the LabChip® automated microfluidic capillary electrophoresis (micro-CE) technology and enable the analysis of genomic DNA and NGS libraries in seconds. Reporting is improved and simplified through the automatic generation of data as an electropherogram, virtual gel, or tabular report.
Suggested links:
16S Amplicon-seq library prep
16S rRNA sequencing are useful and cost-effective NGS tools for microbial sequencing for metagenomic studies comparing different species of bacteria present in a given sample without the need for cultures. 16S rRNA is a widely used gene marker for genus and species identification in bacteria for its minuscule rate of substitution in the highly conserved sequences outside of the hypervariable regions. The speed and economy associated with amplicon sequencing is maximized with this approach catered to population genetics and microbial profiling for interrogating the phylogeny and taxonomy of samples from complex microbiomes.
Suggested products:
Library prep for shotgun sequencing
Shotgun metagenomic sequencing offers researchers a comprehensive approach to sampling all the genes present in a complex sample, encompassing all organisms within it. This method can be invaluable for microbiologists as it allows for the assessment of bacterial diversity and the identification of microbial abundance across diverse environments. Furthermore, shotgun metagenomics facilitates the study of unculturable microorganisms that would otherwise present challenges or even impossibilities in analysis.
We now provide access to the CosmosID-HUB, a leading bioinformatics platform for advanced microbial analysis. The CosmosID-HUB offers comprehensive tools for rapid identification and profiling of microorganisms, helping you interpret complex sequencing data with confidence. This collaboration ensures that users of our NEXTFLEX® Rapid XP V2 DNA-seq kit for Metagenomics have both an optimized library preparation solution and access to state-of-the-art analysis resources.
With a streamlined protocol including fully automated normalization & fragmentation and up to 1,536 UDI Barcodes available for multiplexing, the NEXTFLEX® Rapid XP V2 DNA-seq kit is optimized for shotgun sequencing applications.
Suggested products:
Metatranscriptomics Library prep
Revvity offers library prep solutions for metatranscriptomics including directional RNA-seq library prep kits and CRISPR-Cas9 based ribodepletion solutions to remove uninformative molecules prior to sequencing and improve your ability to detect biologically relevant transcripts in complex communities containing host and bacteria RNA.
Suggested products:
Automation of metagenomics NGS workflows
Revvity NGS liquid handlers’ standardized deck configuration allows for complete liquid handling, thermal control, and waste management for the most complex application requirements in next generation sequencing sample preparation protocols. Revvity's liquid handler portfolio has validated protocols for several metagenomics NGS applications, including shotgun and 16S sequencing.
Suggested links: