
NEXTFLEX Rapid XP V2 DNA-Seq Kit



| Feature | Specification |
|---|---|
| Automation Compatible | Yes |
| Product Group | DNAシーケンス |



Loading...
Product information
Overview
- シングルチューブ酵素断片化、エンドリペア&Aテーリング - 最短2.5時間でDNAをシーケンシング対応ライブラリーへ
- 250 ng以上のサンプルに対応するPCR-freeオプションおよび100 pg - 1 µgの広範囲なサンプル濃度に対応したPCR増幅
- NEXTFLEXのノーマライゼーションビーズ - ライブラリー後の定量化や手動プーリングを省略可能
- 低濃度のアダプターダイマーの形成、および均一なGCカバレッジで確実なバリアント検出
- 大規模な多重化研究に対応する最大1,536 UDI利用可能
- 96 UDI-UMIバーコードセットにより、希少バリアント検出の分子誤差補正が可能
- Revvity Automation NGSワークステーションで検証済みの自動化対応ワークフロー
- 各ロットを徹底的にテストすることで品質を保証しています。
NEXTFLEX Rapid XP V2 DNA-Seq Kit は、速度、データ品質、スケーラビリティを融合しています。酵素断片化によりワークフローが合理化され、独自のノーマライゼーションビーズがQCステップ全体を省略し、オプションのPCRフリー経路がライブラリーの複雑性を保ちます。サンプルを少量処理する場合でも、自動分注機上で96ウェルプレートを処理する場合でも、Illumina®およびElement AVITI™シーケンシングに対応した一貫性のある偏りのないライブラリーを作成します。
Additional product information
One-tube enzymatic fragmentation - libraries in as little as 2.5 hours
The NEXTFLEX enzyme cocktail performs fragmentation, end-repair, and A-tailing in a single tube. A complete manual run, from DNA to normalized libraries, takes as little as 2.5 hours total. Fragment size is tunable by adjusting the 35°C incubation. This streamlined chemistry removes the need for sonication equipment, cuts out an extra cleanup step, and reduces thermocycler runs.
Broad input range WGS with PCR-free option
The protocol accepts 100 pg – 1 µg of genomic DNA. For ≥ 250 ng inputs, users may skip PCR (Appendix A workflow), preserving library complexity and minimizing duplicates; low-input samples use 2–15 cycles as recommended. Expected post-PCR yield is 100–500 ng, sufficient for one or more flow-cell lanes.
Highly Efficient Ligation with Minimal Adapter-Dimer Formation
The NEXTFLEX Rapid XP V2 DNA-Seq Kit delivers exceptional ligation efficiency in a single 20°C incubation.
- 30-minute incubation – keeps total library-prep time at 2.5 hours for time-sensitive runs.
- 60-minute incubation – produces optimal ligation efficiency when the highest possible conversion is critical, for example, PCR-free workflows or ultra-low-input samples.
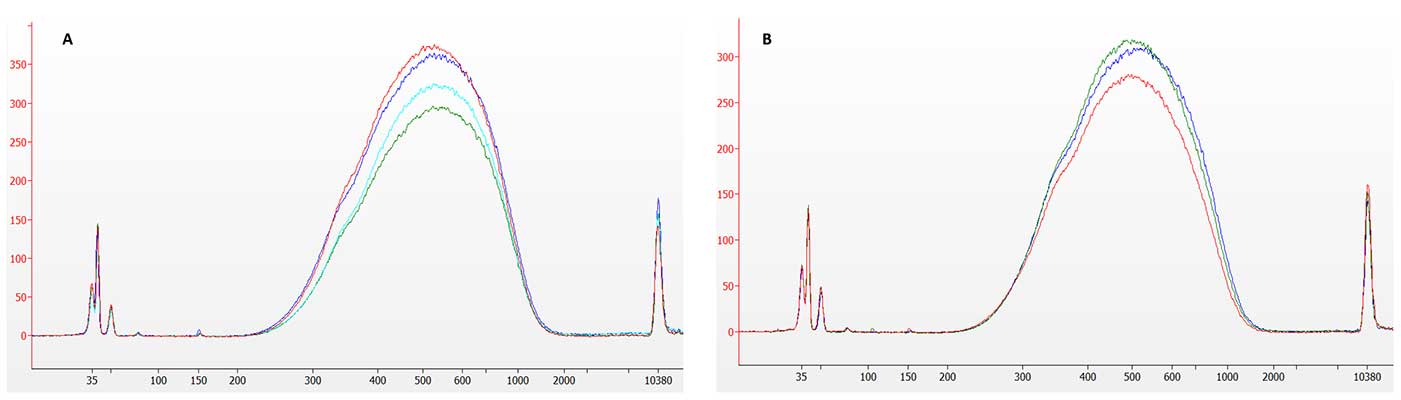
Optimized reagents join adapters without a prior dilution step, converting nearly all fragments across our entire input range. Electropherograms from 500 pg and 1 ng libraries (Figure 1) show no detectable adapter-dimer peak (~150 bp), even at picogram inputs. This combination of high ligation yield and low dimer formation increases usable sequencing reads and ensures reliable, reproducible data.

Figure 1. Electropherogram traces for libraries prepared from 500 pg (A) and 1 ng (B) human gDNA with the NEXTFLEX Rapid XP V2 DNA-Seq Kit. The dominant library peak is flanked by an adapter-dimer region at ~150 bp that is virtually absent, confirming highly efficient ligation and cleanup even at sub-nanogram inputs.
Uniform coverage across the GC spectrum
GC-bias analysis shows flat, normalized depth from ~20% to 60% GC, independent of input mass; only mild drop-off appears above 60%. This consistency yields near-complete genome coverage at standard read depths, boosting SNP/indel sensitivity without expensive over-sequencing.
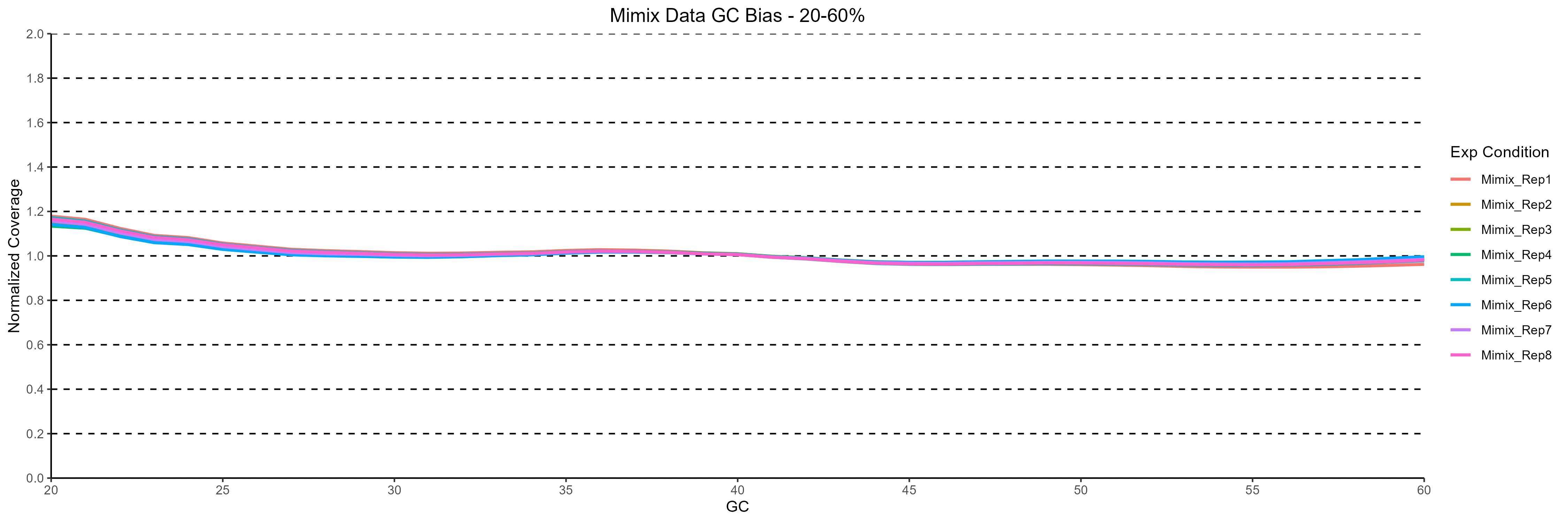
Figure 2. Normalized coverage across varying %GC content for the 8 replicates analyzed. The minimal GC bias observed indicates a high degree of coverage uniformity in the data generated.
Built-in NEXTFLEX Normalization Beads - save ≈ 3 h per 96-sample batch
Proprietary beads bind a fixed mass of library DNA during the final cleanup, so every sample leaves the protocol at the same molarity. This single step simplifies the workflow, reduces hands-on time, and ensures optimal library-input balance for precise pooling and sequencing. By eliminating post-library qPCR/fluorometric quantitation and manual pooling, labs can save up to three hours on a 96-sample run. The on-bead normalization also cuts sample-to-sample variability, preventing costly resequencing due to over- or under-clustering and eliminates the need for quantitation reagents and instrumentation.
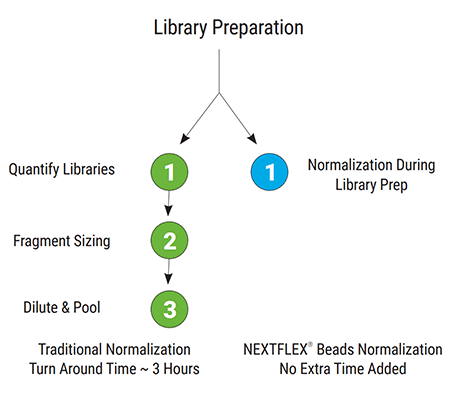
Figure 3. Workflow comparison of traditional post-library normalization versus built-in NEXTFLEX bead normalization. The conventional route requires three downstream QC steps, quantify libraries, fragment sizing, dilute & pooling, adding ~3 hours to turnaround, while NEXTFLEX beads perform normalization during library prep with no extra time.
Indexing options - from duplex-error correction to population-scale multiplexing
NEXTFLEX indexing chemistry is modular, letting you choose the depth of error control and the level of multiplexing your project demands:
| Option | Core benefit | Ideal use-cases |
|---|---|---|
| UDI-UMI plates (10 bp UDI + 9 bp UMI) | Single-molecule quantification & bias-free duplicate removal | cfDNA/liquid-biopsy panels, ultra-deep exome, antimicrobial-resistance screens |
| High-plex UDI sets (8–10 bp UDI) | Massive multiplexing with zero index reuse, perfect for patterned-flow-cell sequencers. | Population-scale WGS, large metagenomic surveys, core-facility batching |
Automation-ready workflows
Revvity provides vendor-qualified scripts for Sciclone™ G3 NGSx/iQ, Zephyr™ NGS, and Fontus™ workstations, so users can drop the NEXTFLEX onto pre-mapped deck positions and run a validated protocol - no custom coding or tip-mapping required.
Specifications
| Automation Compatible |
Yes
|
|---|---|
| Product Group |
DNAシーケンス
|
| Shipping Conditions |
Dual Temperature
|
| Unit Size |
8反応分
|
Citations
- Gaulke, C. A., Schmeltzer, E. R., Dasenko, M., et al. (2021). Evaluation of library preparation procedure and sample characteristics on metagenomic profiles. mSystems, 6(5), e00440-21.
- Author(s). (2024). Beyond single diagnosis: Exploring multidiagnostic realities in [Journal Name]. https://doi.org/10.1155/2024/9115364
- Di Martino M L, Jenniches L, Bhetwal A, Eriksson J, Lopes A C C, Ntokaki A, Pasqua M, Sundbom M, Skogar M, Graf W, Webb D-L, Hellström P M, Mateus A, Barquist L, & Sellin M E. (2025). A scalable gut epithelial organoid model reveals the genome-wide colonization landscape of a human-adapted pathogen. Nature Genetics. Advance online publication. https://www.nature.com/articles/s41588-025-02218-x
FAQs
-
How long does library prep take with the NEXTFLEX Rapid XP DNA-seq kit v2?
-
Do I still need mechanical shearing or tagmentation?
-
What DNA input range is supported, and when can I skip PCR?
-
How do Normalization Beads save time?
-
How uniform is coverage across different GC contents?
-
What level of adapter-dimer formation should I expect?
-
Which sequencing platforms are compatible?
-
Do I still use Normalization Beads if follow PCR-free Protocol?
-
What does Revvity do to assure quality?
Resources
Are you looking for resources, click on the resource type to explore further.
Read the Mimix brochure to explore our range of cell line-derived Mimix reference standards which provide more patient-like...
The new NEXTFLEX® Rapid XP V2 DNA-seq kit includes proprietary NEXTFLEX® normalization beads*, which provide a consistent mass and...
Streamline WGS library prep for newborn sequencing applications with NEXTFLEX Rapid XP v2 to enable enhanced variant detection for...
Loading...
Loading...


How can we help you?
We are here to answer your questions.






























