Analyzing circulating cell free DNA (ccfDNA) offers a non-invasive, dynamic window into the body’s physiological state and its use as a biomarker is gaining rapidly. However, multiple challenges remain with establishing its routine application. This is mainly due to its low levels, high rates of fragmentation, and preanalytical variabilities associated with sample collection, cfDNA extraction and quantification. Here, we go through some tips on how one can establish a reliable cfDNA workflow from plasma samples.
1. Sample Collection
Cell free DNA is found in various body fluids and can be shed from cells undergoing apotosis or necrosis, or actively released from live cells1. Plasma is the most well-studied sample type and is recommended over serum as the latter tends to experience higher genomic DNA contamination attributed to white blood cell (WBC) lysis during the clotting process2. Depending on the application, sample volume required should be established beforehand. Striving to detect low frequency mutations in circulating tumour DNA (ctDNA) for example, may require higher plasma input volumes which may necessitate drawing a higher volume of blood.
To minimize genomic DNA contamination when isolating cfDNA from plasma,
- Minimize cell lysis during blood drawing e.g. use appropriate needle size, avoid prolonged tourniquet
- Avoid harsh temperature changes and excessive agitation when storing or transporting blood tubes
- Isolate plasma carefully, avoiding contact with buffy coat layer (learn how you can automate this with Revvity’s JANUS G3 Blood IQ Workstation
- Isolate plasma within 6 h of blood collection when using EDTA blood tubes3, or longer for specialized cell free blood collection tubes containing stabilizers (refer to tube manufacturer’s instructions)
- Perform a double centrifugation of plasma to minimize carryover of WBCs
- Store isolated plasma at -80°C if not using immediately and avoid freeze thaw cycles
2. Extraction of cfDNA from Plasma
Extraction of cfDNA typically relies on silica columns or magnetic beads to concentrate cfDNA from large plasma volumes, followed by several wash steps and elution in a smaller buffer volume for downstream processing. There have been studies comparing commercially available kits which report varying yields and quality4. However, these results are also influenced by variabilities in sample source, handling steps and quantification procedures across labs.
We tested the chemagic™ cfDNA extraction kits5, based on patented M-PVA Magnetic Beads, on the chemagic 360 instrument and obtained comparable and consistent yields to a competitor’s manual silica column and vacuum-based method.
As different extraction kits have reportedly variable extraction efficiencies based on DNA fragment length, we also analyzed fragmentation of obtained cfDNA by PCR using the KAPA™ Human Genomic DNA Quantification and QC Kit from Roche. Fragmentation/degradation scores with the chemagic extraction kits were in the range of (0.16 - 0.37) which was comparable to the results gained with the silica membrane-based competitor kit on the same samples (degradation scores 0.15 - 0.4).

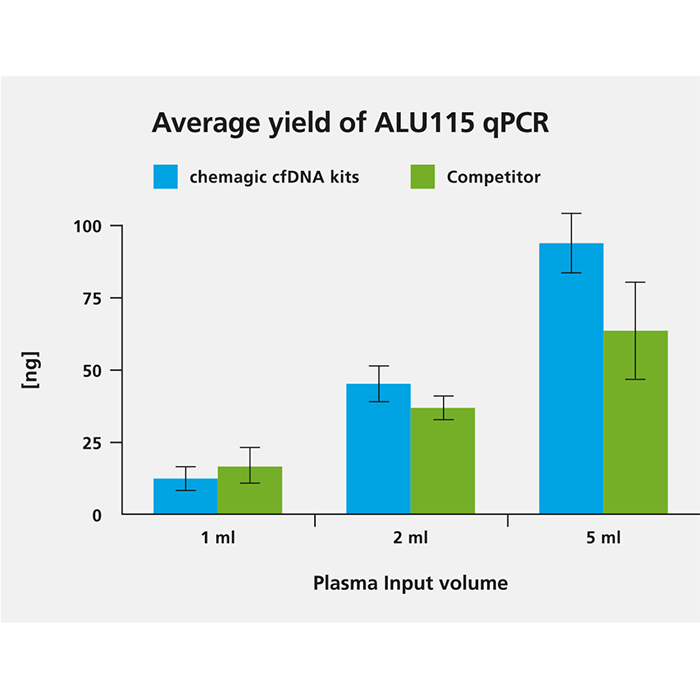
cfDNA was isolated from 1 ml, 2 ml and 5 ml from two donors both with the chemagic kits on the chemagic 360 instrument and manually with competitor. For cfDNA analysis, a short fragment (115 bp) from a consensus sequence with abundant genomic ALU repeats was amplified. Exemplary data from donor 2 shows that the yield of cfDNA is scalable to sample input.
By automating cfDNA extraction with chemagic™ technology, one can:
- Increase sample throughput while maintaining comparable yields to manual methods
- Reduce hands-on steps and speed up workflow (< 2 h for up to 96 samples with on-board lysis and no heating required)
- Enable a wider range of sample inputs (0.5 to 18 ml applicable)
- Maintain sample integrity with sample tracking options (barcode reading and bidirectional LIMS communication capability)
- Reduce running cost per sample
- Increase downstream success with various assays such as digital droplet PCR (ddPCR), Next-Generation Sequencing (NGS), quantitative PCR (qPCR) etc. (see references)
As cfDNA levels vary widely between samples, one may also include an exogenous DNA control in samples to monitor extraction efficiency6.
3. Quantification and Quality Control of Extracted cfDNA
Before cfDNA is subject to downstream sequencing, quality control checks are often performed to evaluate the quality of extracted cfDNA. As cfDNA yields isolated from human plasma samples are typically critically low in the range of 1 - 30 ng/mL of plasma, this may lie outside the detection parameters determined by spectrophotometric methods. Hence, if quantification of the extracted cfDNA is required, a PCR-based method (e.g. qPCR or ddPCR) is recommended.
When using fluorometric quantification methods, the addition of Poly(A) RNA is essential for reliable performance. Fluorometric analysis of eluates extracted without the use of Poly(A) RNA may lead to varying and decreased quantification data. Additionally, care should be taken in using fluorometric methods for quantification as not only cfDNA, but total DNA is measured.
To assess fragment distribution of the extracted cfDNA, fragment analysis systems, such as LabChip™ GX Touch™ HT Nucleic Acid Analyzer from Revvity, may be used. A major peak at around 150 - 170 bp is expected for high quality mononucleosomal cfDNA, in some cases a smaller peak at around 300 bp representing dinucleosomal cfDNA fragments is also present. The use of fragment analysis systems for cfDNA quantification did not lead to reproducible results during our kit development and is not recommended. Marker peak areas (marker included in fragment analyzer kits) vary according to different extraction chemistries. Variations of marker peak areas within one fragment analysis run may result in miscalculations.
Our recommended cfDNA quantitation procedure involves:
- qPCR using an ALU115 primer set where a short fragment (115 bp) from a consensus sequence with abundant genomic ALU repeats was amplified
- other qPCR/ddPCR-based assays targeting “housekeeping” genes found in cfDNA which have been experimentally validated7
- fluorometric/fragment analyzer approaches may be used for qualitative assessment of cfDNA but may lead to variable quantification results
Extracted cfDNA from chemagic cfDNA extraction kits have been successfully put through library preparation with the NEXTFLEX Cell Free DNA-Seq Library Prep Kit 2.0. The option to automate this process can further contribute to a reliable cfDNA workflow by minimizing risk of errors while increasing operational capacity8.
Discover our Solutions for cfDNA & cfRNA workflows
https://www.revvity.com/de-en/category/cell-free-dna-analysis
Revvity is a trademark of Revvity, Inc. All other trademarks are the property of their respective owners.
For research use only. Not for use in diagnostic procedures.
References
- van der Vaart, M., & Pretorius, P. J. (2007). The Origin of Circulating Free DNA. Clinical Chemistry, 53(12), 2215–2215. https://doi.org/10.1373/CLINCHEM.2007.092734
- Wong, F. C. K., Sun, K., Jiang, P., Cheng, Y. K. Y., Chan, K. C. A., Leung, T. Y., Chiu, R. W. K., & Lo, Y. M. D. (2016). Cell-free DNA in maternal plasma and serum: A comparison of quantity, quality and tissue origin using genomic and epigenomic approaches. Clinical Biochemistry, 49(18), 1379–1386. https://doi.org/10.1016/J.CLINBIOCHEM.2016.09.009
- Merker, J. D., Oxnard, G. R., Compton, C., Diehn, M., Hurley, P., Lazar, A. J., Lindeman, N., Lockwood, C. M., Rai, A. J., Schilsky, R. L., Tsimberidou, A. M., Vasalos, P., Billman, B. L., Oliver, T. K., Bruinooge, S. S., Hayes, D. F., & Turner, N. C. (2018). Circulating tumor DNA analysis in patients with cancer: American society of clinical oncology and college of American pathologists joint review. Journal of Clinical Oncology, 36(16), 1631–1641. https://doi.org/10.1200/JCO.2017.76.8671
- Lampignano, R., Consortium, for the I. M. I. C.-I., Neumann, M. H. D., Consortium, for the I. M. I. C.-I., Weber, S., Consortium, for the I. M. I. C.-I., Kloten, V., Consortium, for the I. M. I. C.-I., Herdean, A., Consortium, for the I. M. I. C.-I., Voss, T., Consortium, for the I. M. I. C.-I., Groelz, D., Consortium, for the I. M. I. C.-I., Babayan, A., Consortium, for the I. M. I. C.-I., Tibbesma, M., Consortium, for the I. M. I. C.-I., Schlumpberger, M., … Consortium, for the I. M. I. C.-I. (2020). Multicenter Evaluation of Circulating Cell-Free DNA Extraction and Downstream Analyses for the Development of Standardized (Pre)analytical Workflows. Clinical Chemistry, 66(1), 149–160. https://doi.org/10.1373/CLINCHEM.2019.306837
- Technical Note: Automated Circulating Cell-Free DNA Purification with the chemagic™ 360 Instrument.
- Pallisgaard, N., Spindler, K. L. G., Andersen, R. F., Brandslund, I., & Jakobsen, A. (2015). Controls to validate plasma samples for cell free DNA quantification. Clinica Chimica Acta, 446, 141–146. https://doi.org/10.1016/J.CCA.2015.04.015
- Lefèvre, A. C., Pallisgaard, N., Kronborg, C., Wind, K. L., Krag, S. R. P., & Spindler, K. L. G. (2021). The clinical value of measuring circulating hpv dna during chemo-radiotherapy in squamous cell carcinoma of the anus. Cancers, 13(10). https://doi.org/10.3390/cancers13102451
- Application Note: High-Throughput, End-to-End Cell Free DNA Analysis Workflow from Plasma.