RNA sequencing library prep kits
RNA sequencing (RNA-seq) is used to analyze the quantity, and the sequence of all RNA transcripts present in a sample at any given time, which is often referred as transcriptome. The transcriptome includes all types of RNA, including messenger RNA (mRNA), ribosomal RNA (rRNA), transfer RNA (tRNA) and various non-coding RNAs (ncRNAs). The transcriptome represents the genes that are actively being expressed in a cell or tissue, providing a snapshot of cellular function and activity.
The main applications where RNA-seq is used are listed below:
Gene Expression Profiling: Quantifies the expression levels of mRNA in a sample, providing insights into which genes are active.
Differential Expression Analysis: Closely related to previous application, but in this case the focus is on detecting genes that are upregulated or downregulated in response to specific conditions, such as comparing healthy versus diseased tissues.
Transcriptome Assembly: Constructs a comprehensive map of all RNA transcripts, revealing alternative splicing events, novel transcripts, and gene structures, .
Non-Coding RNA Identification: Identifies and studies the regulatory role of various ncRNAs, such as microRNAs, long non-coding RNAs and circular RNAs.
Revvity provides a full portfolio of modular products and accessories designed to facilitate RNA-seq for customers utilizing Illumina & Element Biosciences sequencing instruments. Please continue reading to understand what portions of our portfolio may be of use to your lab.
For research use only. Not for use in diagnostic procedures.
RNA Sequencing Library Prep
RNA cannot be sequenced directly. Therefore, library preparation involves converting RNA into complementary DNA (cDNA) using retrotranscriptase, followed by second strand synthesis and addition of barcoded adapters. Most workflows have introduced modifications to generate directional libraries, which allow determining which DNA strand the RNA transcript originated from. This property can be relevant when analyzing complex genomes.
There are two strategies commonly used for cDNA synthesis that are worth discussing. The first one utilizes oligo (dT) primers that bind to the polyadenylated tails of the 3’ end of mRNA. It is ideal for mRNA but not suitable for non-polyadenylated RNA species or gene expression studies on prokaryotes. It is often restricted to samples of high integrity, because when there is degradation information from 5’ end of the transcript may be lost. The second strategy is more versatile and is based on the usage of random primers that bind through the RNA.
This provides a more uniform coverage across the entire length of the transcript than previous approach and can be used to study any RNA species. For gene expression profiling studies, where the interest is only mRNA, additional enrichment or depletion steps are required when using this strategy.
Revvity’s workflow generates directional libraries using random primers for cDNA synthesis. We also provides accessories for enrichment and depletion, that can be implemented before, during and after library preparation, depending of the option chosen (see Selection Guide below).Standard UDI can be used but for applications where precise copy number quantitation is required, UDI-UMIs are preferred as they allowed to remove PCR duplicates (Figure 1).
For RNA molecules with a size of 100 nucleotides or less conversion of RNA into cDNA can be challenging. In some extreme cases such as microRNA, the length of the molecule of RNA we want to sequence (19-25 nt) has a size comparable to a primer. To overcome this problem a different approach is needed, where the targets of interest are tagged with special adapters that will provide a priming site for cDNA synthesis downstream. Revvity offers a dedicated solution for these studies, the NEXTFLEX Small RNA Sequencing kit v4.
Enrichment
Bead-based poly(A) enrichment is commonly used to isolate mRNA from total RNA. Magnetic beads coated with oligo(dT) sequences are used to selectively hybridize to the poly(A) tail found at the 3’ end of most eukaryotic mRNAs. After binding, the beads are washed to remove non-target RNA, and the mRNA is then eluted from the beads. This method provides a high-purity mRNA sample that can be used as input for Revvity or any other RNA-seq library preparation workflow downstream.
Depletion
Depletion (also known as ribodepletion), focuses on reducing the presence of highly abundant RNA species. There are different approaches available.
A popular method is bead-based rRNA depletion, where like in the previous case magnetic beads are used to selective bind rRNA, which is then discarded. By depleting rRNA, that constitutes 80-90% of the total RNA, researchers can enhance the detection of low-abundance transcripts. This method is preferred when RNA is degraded, or when we are interested in RNA species that may not be polyadenylated, such as long non-coding RNAs.
CRISPR-based depletion is an innovative technique used to remove unwanted sequences from a sample. This method utilizes the CRISPR/Cas9 system, where guide RNAs are designed to target specific dsDNA for cleavage by the Cas9 enzyme. By creating double-stranded breaks at these targeted sites, the unwanted sequences are then removed. CRISPR-based depletion can be used with low input amounts (lower than bead-based depletion), and can be applied within or after library preparation. Revvity offers depletion solutions based on magnetic beads and CRISPR/Cas9 system. For a deeper dive into ribodepletion, check out our Ribodepletion Solutions.
Selection Guide:
| Workflow Step | Multiplexing | Sequencers | Revvity Solution |
|---|---|---|---|
| Poly(A) Enrichment | RNA-seq barcodes or UDI-UMI barcodes | Illumina® Element® Platforms | NEXTFLEX Poly(A) Beads 2.0 |
| Ribodepletion | Ribodepletion Solutions | ||
| RNA Library Prep | NEXTFLEX Rapid Directional RNA-Seq Kit 2.0 |
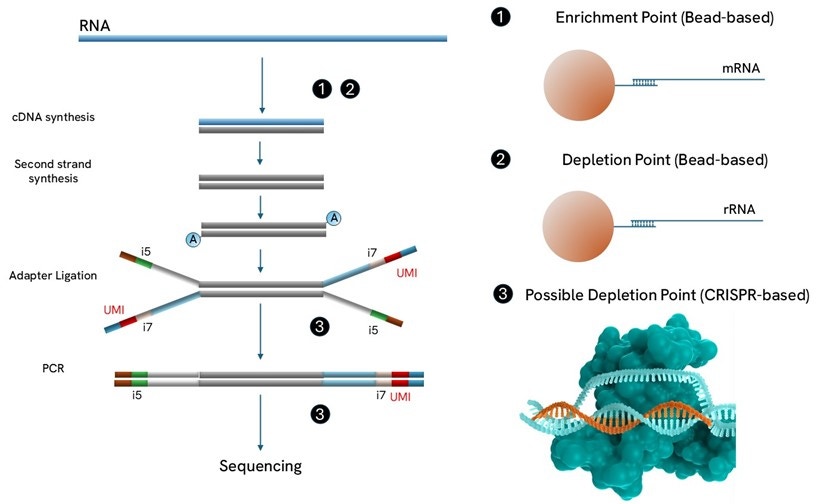
Figure 1. This figure illustrates the classic RNA-seq workflow. The numbers in the diagram show where various enrichment & depletion based accessory products can be used in RNA-seq workflows.





























