SPRI technology and the impact of bead-to-sample ratio
Solid Phase Reversible Immobilization (SPRI) beads were originally developed at the Whitehead Institute in 1995 for nucleic acid purification. Since that time, they have become crucial component in various applications. Solid Phase Reversible Immobilization (SPRI) beads were originally developed at the Whitehead Institute in 1995 for nucleic acid purification. Since that time, they have become crucial component in various applications, — from SPRI cleanup steps in next-generation sequencing (NGS) library preparation to PCR clean-up and plasmid purification.
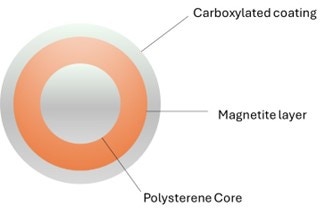
SPRI beads are paramagnetic particles (magnetic only in a magnetic field) of a few micrometers of diameter. They are made of a core of polystyrene surrounded by a layer of magnetite, responsible of their characteristic brown color. The magnetite layer is coated with carboxyl molecules that reversibly bind DNA or RNA under specific conditions (Figure 1).

Figure 1. Section of a typical SPRI bead
How do SPRI beads work
The key principle behind SPRI technology lies in its ability to bind nucleic acids in the presence of the crowding agent polyethylene glycol (PEG) and salts. PEG attracts and binds water molecules, reducing the solubility of the DNA and their volume available in solution. The presence of salts can further enhance precipitation by neutralizing the charges on the nucleic acid molecules, which reduces the repulsion between them and favor interaction with the carboxyl groups. This binding is reversible, allowing for easy elution of the nucleic acids using water or low-salt buffers.
The concentration of PEG in solution is critical in the concentration of PEG in solution is critical in SPRI beads size-selection, because higher-molecular-weight DNA precipitates at lower PEG concentrations than low-molecular-weight DNA. In other words, the size of the fragments that bind to the SPRI beads is determined by the concentration of PEG, and this is in turn determined by the volume of beads and DNA.
A mix of 25 μL SPRI beads and 25 μL of DNA will give a bead-to-sample ratio of 1x, whereas a mix of 25 μL SPRI beads and 50 μL of DNA will give a ratio of 0.5x.
With low bead-to-sample ratio, only large fragments bind to the beads; smaller fragments remain in solution and can be discarded — an efficient form of SPRI cleanup for removing adapter dimers during NGS library prep. This property is particularly useful in NGS library preparation, where specific size ranges are often desired for optimal sequencing performance. This property is particularly useful in NGS library preparation, where specific size ranges are often desired for optimal sequencing performance. For example, keeping a low bead-to-sample ratio (typically ~0.8x), researchers can effectively remove adapter dimers and retain larger fragments for sequencing. Higher bead-to-sample ratios tend to bind all DNA molecules, including smaller fragments (Figure 2). Higher ratios tend to increase yield obtained.
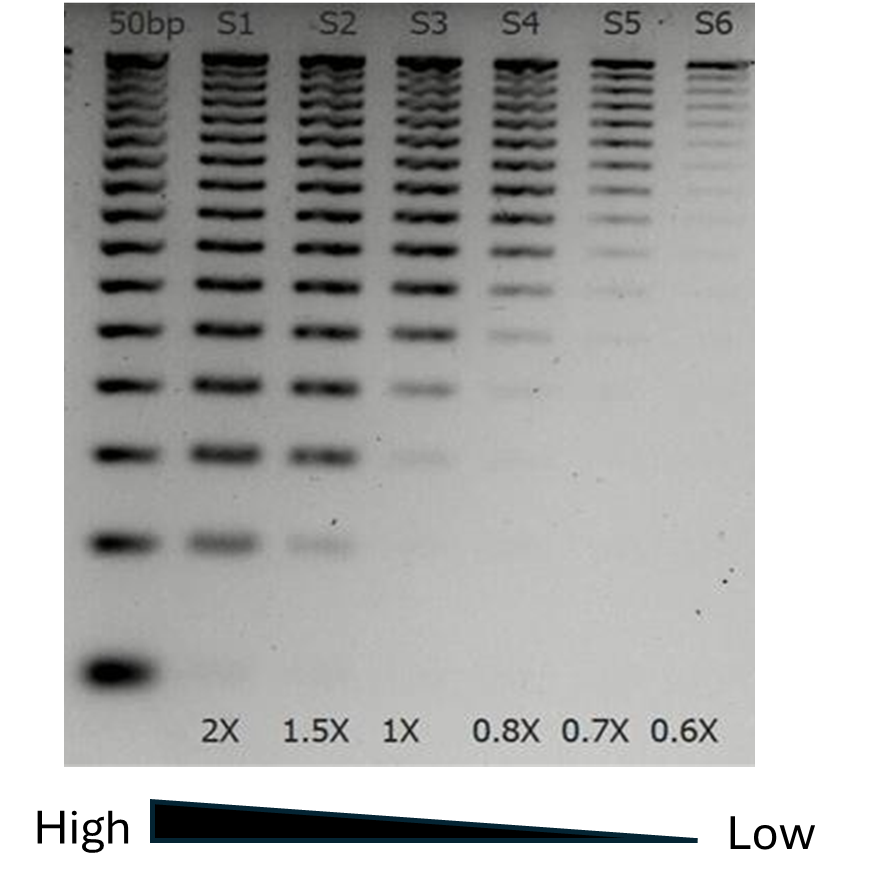
Figure 2. Impact of bead-to-sample ratio in size selection. A fixed amount of 50 bp DNA ladder (Promega) was purified with different volumes of NEXTFLEX™ NGS Cleanup beads
 NEXTFLEX NGS Cleanup Beads
Discover
. Eluted DNA was run on 1% agarose gel. It can be clearly seen that at low ratios only large molecules bind to the beads, while smaller fragments remain in solution.
NEXTFLEX NGS Cleanup Beads
Discover
. Eluted DNA was run on 1% agarose gel. It can be clearly seen that at low ratios only large molecules bind to the beads, while smaller fragments remain in solution.
In practice, researchers often need to optimize the bead-to-sample ratio for their specific applications. This may involve testing a range of ratios to find the best compromise between yield, purity, and size selection. Some protocols employ multiple rounds of SPRI purification with different ratios to achieve tighter size selection and purity.
For research use only. Not for use in diagnostic procedures.


































