
NEXTFLEX UDI Barcodes (10NT, 769-1152)

NEXTFLEX UDI Barcodes (10NT, 769-1152)




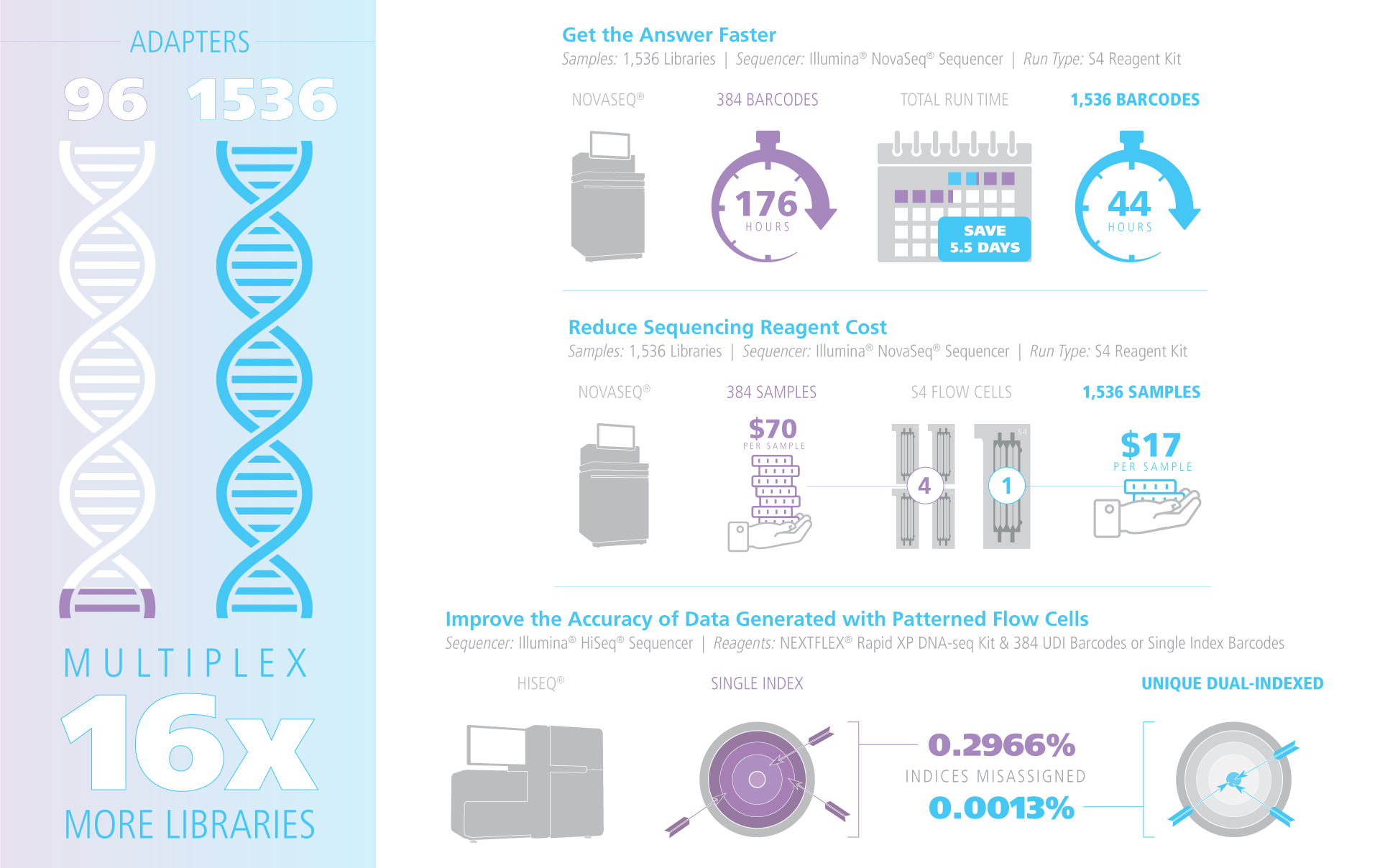
Multiplex up to 1,536 samples on a single flow cell with NEXTFLEX™ Unique Dual Index Barcodes (10NT UDIs). Each kit ships with up to 1,536 pre-validated i5/i7 index pairs that eliminate index hopping and are compatible with Illumina platforms such as the NovaSeq® and Element’s AVITI™ systems. Off-the-shelf plate sets (containing between 16 to 1,536 unique adapters) let you start small and scale to population-level projects without requiring custom ordering or re-balancing.
| Feature | Specification |
|---|---|
| Automation Compatible | Yes |
| Product Group | Barcodes |
Multiplex up to 1,536 samples on a single flow cell with NEXTFLEX™ Unique Dual Index Barcodes (10NT UDIs). Each kit ships with up to 1,536 pre-validated i5/i7 index pairs that eliminate index hopping and are compatible with Illumina platforms such as the NovaSeq® and Element’s AVITI™ systems. Off-the-shelf plate sets (containing between 16 to 1,536 unique adapters) let you start small and scale to population-level projects without requiring custom ordering or re-balancing.


NEXTFLEX UDI Barcodes (10NT, 769-1152)


NEXTFLEX UDI Barcodes (10NT, 769-1152)


Loading...
Product information
Overview
- Up to 1,536 unique i5 / i7 pairs (10NT) ready to ship
- Compatible with PCR-free and PCR-amplified workflows
- ≥ 3-base Hamming distance across the entire set for robust error-correction and balanced base diversity on 2- & 4-color chemistries
- Designed to minimize index hopping that can be present with patterned flow cells.
- Compatible with Revvity library prep and TruSeq style, ligation-based workflows
- Every lot is functionally validated and tested for index purity by sequencing in an ISO 9001 facility
- Optional NEXTFLEX Universal Blockers boost on-target hybrid-capture efficiency when used with these UDIs.
Additional product information
NGS Multiplexing Performance
Scale from 2 to 1,536 libraries in a single NovaSeq® (Illumina) or AVITI® (Element)flow cell. The 10NT UDI set supports barcode rotation between runs, eliminating carry-over and virtually stopping index hopping on patterned flow cells. Adapters work out-of-the-box with all TruSeq-style library-prep kits, including NEXTFLEX, Illumina, Watchmaker, Twist library prep kit s, whether PCR-free or PCR-amplified.
“Revvity has a set of 1536 validated UDI adapters that we have been regularly using for 5 years. Using the Illumina® NovaSeq® X Plus with this UDI set from Revvity we can sequencing 12,288 samples across 8 lanes of the 10B or 25B flow cells. This dramatically lowers our sequencing cost per sample!”
-Dr. Charlie Johnson, Director of Texas A&M Agri Life Genomics and Bioinformatics Service
NGS Adapters with High Data Integrity
Each NEXTFLEX UDI adapter has a dedicated 10NT i7 & i5 sequence separated from every other pair by a ≥ 3-base Hamming distance, enabling single-base error correction during demultiplexing. Because the adapter already contains the full flow-cell and primer-binding sites, PCR enrichment is optional. On patterned flow cells (Illumina NovaSeq®, HiSeq® 3000/4000, HiSeq® X, Element AVITI™) the use of true, unique dual indexes cuts index-hopping artefacts to background levels by removing mis-assigned reads. Every lot is sequenced for ≥ 99.9 % index purity and functional ligation efficiency, ensuring data integrity run after run.

Specifications
| Automation Compatible |
Yes
|
|---|---|
| Barcodes |
769 - 1152
|
| Format |
2 rxns per well
|
| Product Group |
Barcodes
|
| Shipping Conditions |
Shipped in Dry Ice
|
| Unit Size |
768 rxns
|
References
Selected Citations that Reference the Use of the NEXTFLEX UDI Barcodes:
- Adolfi, A., Gantz, V.M., Jasinskiene, N. et al. Efficient population modification gene-drive rescue system in the malaria mosquito Anopheles stephensi. Nat Commun 11, 5553 (2020).
- Estermann, M. (2020). Mouse embryonic stem cells self-organize into trunk-like structures with neural tube and somites. doi:10.1242/prelights.18906.
- Gardner, E. J., Prigmore, E., Gallone, G., Danecek, P., Samocha, K. E., Handsaker, J., . . . Hurles, M. E. (2019). Contribution of retrotransposition to developmental disorders. Nature Communications, 10(1). doi:10.1038/s41467-019-12520-y.
- Gaeta, N. C., Bean, E., Miles, A. M., Daniel Ubriaco Oliveira Gonçalves De Carvalho, Alemán, M. A., Carvalho, J. S., . . . Ganda, E. (2020). A Cross-Sectional Study of Dairy Cattle Metagenomes Reveals Increased Antimicrobial Resistance in Animals Farmed in a Heavy Metal Contaminated Environment. Frontiers in Microbiology, 11. doi:10.3389/fmicb.2020.590325.
- Hirose, K., Chang, S., Yu, H., Wang, J., Barca, E., Chen, X., . . . Huang, G. N. (2019). Loss of a novel striated muscle-enriched mitochondrial protein Coq10a enhances postnatal cardiac hypertrophic growth. doi:10.1101/755793.
- Leon, K. E., et al. (2020) DOT1L modulates the senescence-associated secretory phenotype through epigenetic regulation of IL1A. bioRxiv 2020.08.21.258020; doi: 10.1101/2020.08.21.258020
- Miura, H., Takahashi, S., Shibata, T. et al. Mapping replication timing domains genome wide in single mammalian cells with single-cell DNA replication sequencing. Nat Protoc 15, 4058–4100 (2020). doi.org/10.1038/s41596-020-0378-5
- Starr, T. N., Greaney, A. J., Hilton, S. K., Crawford, K. H., Navarro, M. J., Bowen, J. E., Bloom, J. D. (2020). Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. doi:10.1101/2020.06.17.157982.
- Veenvliet, J. V., et al. (2020) Mouse embryonic stem cells self-organize into trunk-like structures with neural tube and somites. bioRxiv 2020.03.04.974949; doi.org/10.1101/2020.03.04.974949.
- Dubey SK et al. (2025). Deciphering age-related transcriptomic changes in the mouse retinal pigment epithelium. Aging (Albany NY) 17(3):657–684. DOI: 10.18632/aging.206219
- Pujari & Cullen (2024) Modulators of MAPK pathway activity during filamentous growth in Saccharomyces cerevisiae. G3 14(6):jkae072
- Kant et al. (2023) High-quality full genome assembly of historic Xylella fastidiosa strains… Microbiol Resour Announc. 12(11):e00536-23.
- Arras et al. (2023) Characterisation of an E. coli line that completely lacks ribonucleotide reduction… eLife 12:e83845.
- Larkin et al. (2025) Climate-driven succession in marine microbiome biodiversity and biogeochemical function. Nat Commun. 16:3926.
- Klauer et al. (2024) Hydrophobins from Aspergillus mediate fungal interactions with microplastics. bioRxiv preprint 2024.11.05.622132.
FAQs
-
Are the UDI barcodes sequence-verified and functionally validated?
-
What is the lead time when I order the NEXTFLEX unique dual index barcodes?
-
Must I perform paired-end sequencing with NEXTFLEX Unique Dual Index Barcodes?
-
Can I use the NEXTFLEX Unique Dual Index Barcodes in single index experiments?
-
What are the storage and shipping conditions?
-
Do the UDIs work with PCR-free workflows?
-
Can I combine just a subset of the indexes in one run?
-
How do unique dual indexes mitigate index hopping?
Resources
Are you looking for resources, click on the resource type to explore further.
This application note describes the benefits of using UDI Barcodes.
This flyer describes the benefits of the NEXTFLEX® 1536 UDI Barcode Adapter Set.


How can we help you?
We are here to answer your questions.






























