
HTRF Human Total SALL4 Detection Kit, 10,000 Assay Points

HTRF Human Total SALL4 Detection Kit, 10,000 Assay Points




This HTRF kit allows for enable the cell-based quantitative detection of Total SALL4.
| Feature | Specification |
|---|---|
| Application | Protein Quantification |
| Sample Volume | 16 µL |
This HTRF kit allows for enable the cell-based quantitative detection of Total SALL4.


HTRF Human Total SALL4 Detection Kit, 10,000 Assay Points


HTRF Human Total SALL4 Detection Kit, 10,000 Assay Points


Product information
Overview
SALL4 is a transcription factor from the spalt-like (SALL) gene family, known for its zinc finger domains and crucial role in maintaining the pluripotency and self-renewal of embryonic stem cells. Although SALL4 expression decreases significantly in most adult tissues, it is aberrantly reactivated in various cancers, promoting cell proliferation and stem cell-like characteristics in malignant cells. This re-expression links SALL4 to tumorigenesis and poor prognosis, making it a notable biomarker and potential therapeutic target in oncology.
Immunomodulatory imide drugs (IMIDs), including lenalidomide and thalidomide derivatives, have shown promise in targeting SALL4, especially the SALL4A isoform. These drugs have demonstrated anti-tumor activity but pose a significant risk due to their teratogenic effects, causing severe birth defects. Despite this limitation, SALL4 remains an attractive target in aggressive cancers where its expression is aberrant, especially when other treatments have failed. Ongoing research aims to develop more selective and less toxic inhibitors, balancing the therapeutic benefits of targeting SALL4 with the risks of teratogenicity, thus offering new potential in cancer treatment.
Specifications
| Application |
Protein Quantification
|
|---|---|
| Brand |
HTRF
|
| Buffer/Solvent |
Lysis Buffer 4
|
| Detection Modality |
HTRF
|
| Host Species |
Human
|
| Molecular Modification |
Total
|
| Product Group |
Kit
|
| Sample Volume |
16 µL
|
| Shipping Conditions |
Shipped in Dry Ice
|
| Target |
SALL4
|
| Target Class |
Phosphoproteins
|
| Target Species |
Human
|
| Technology |
TR-FRET
|
| Therapeutic Area |
Inflammation
Oncology
|
| Unit Size |
10,000 assay points
|
How it works
Total SALL4 assay principle
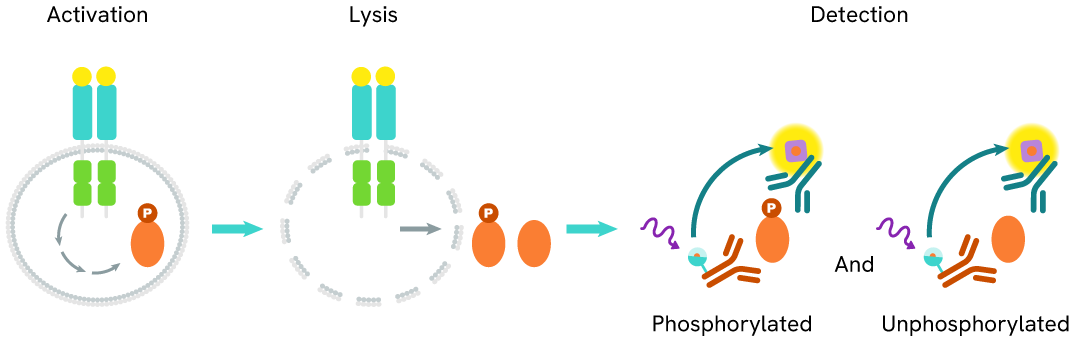
The Total SALL4 assay quantifies the expression level of SALL4 in a cell lysate. Unlike Western Blot, the assay is entirely plate-based and does not require gels, electrophoresis, or transfer. The Total SALL4 assay uses two labeled antibodies, one coupled to a donor fluorophore and the other to an acceptor. Both antibodies are highly specific for a distinct epitope on the protein. In the presence of SALL4 in a cell extract, the addition of these conjugates brings the donor fluorophore into close proximity with the acceptor and thereby generates a FRET signal. Its intensity is directly proportional to the concentration of the protein present in the sample and provides a means of assessing the protein's expression under a no-wash assay format.

Total SALL4 two-plate assay protocol
The two-plate protocol involves culturing cells in a 96-well plate before lysis, then transferring lysates into a 384-well low volume detection plate before the addition of HTRF Total SALL4 detection reagents. This protocol allows the cells' viability and confluence to be monitored.
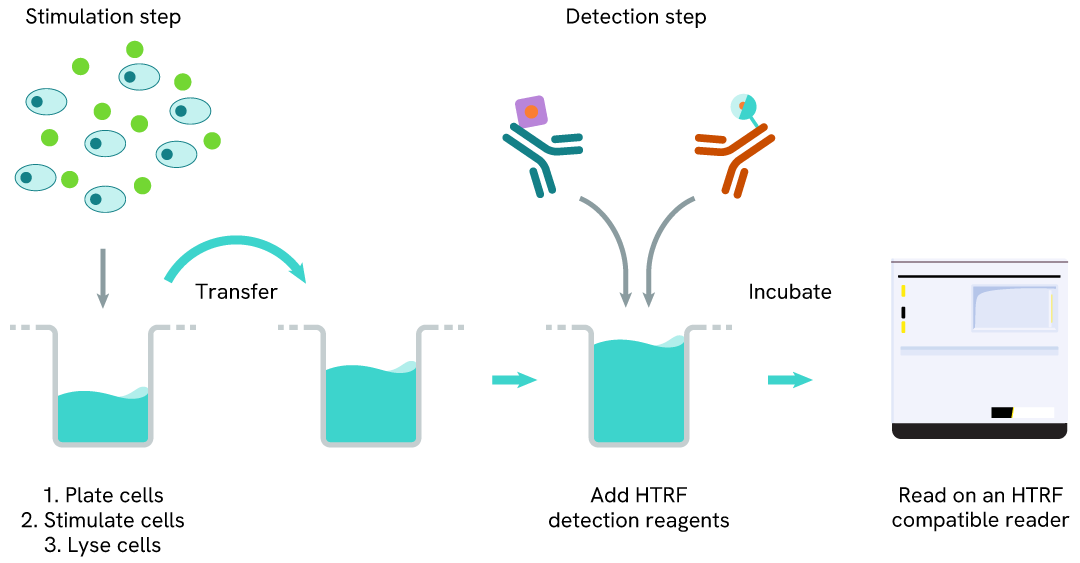
Total SALL4 one-plate assay protocol
Detection of Total SALL4 with HTRF reagents can be performed in a single plate used for culturing, stimulation, and lysis. No washing steps are required. This HTS designed protocol allows for miniaturization while maintaining robust HTRF quality.
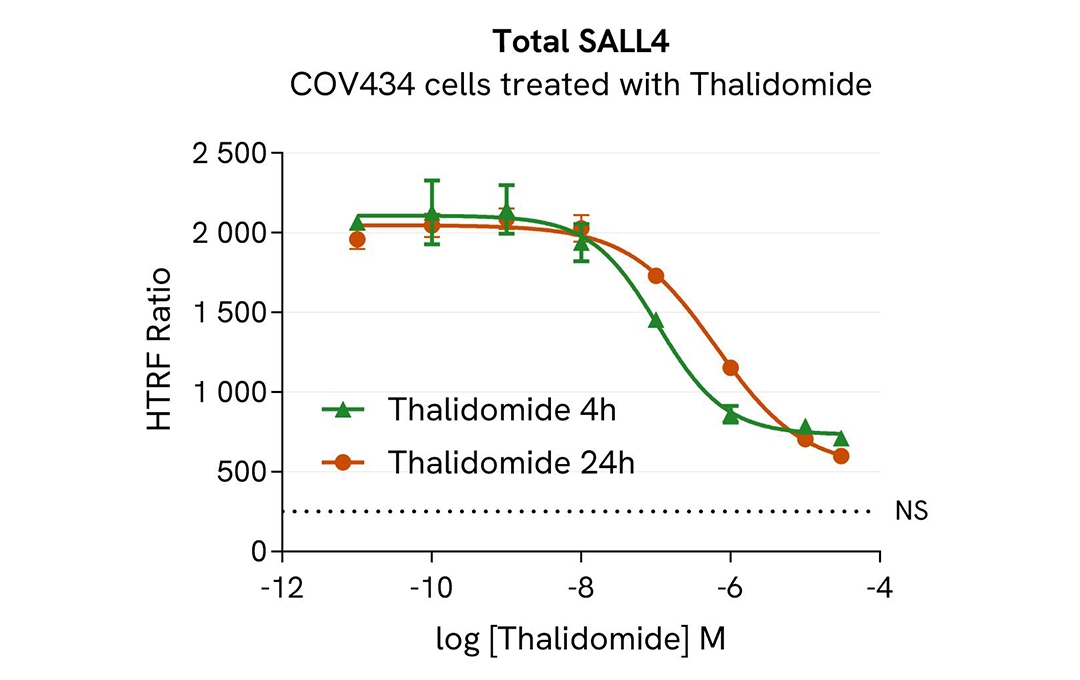
Assay validation
HTRF Total SALL4 modulation using Thalidomide on COV434 cells
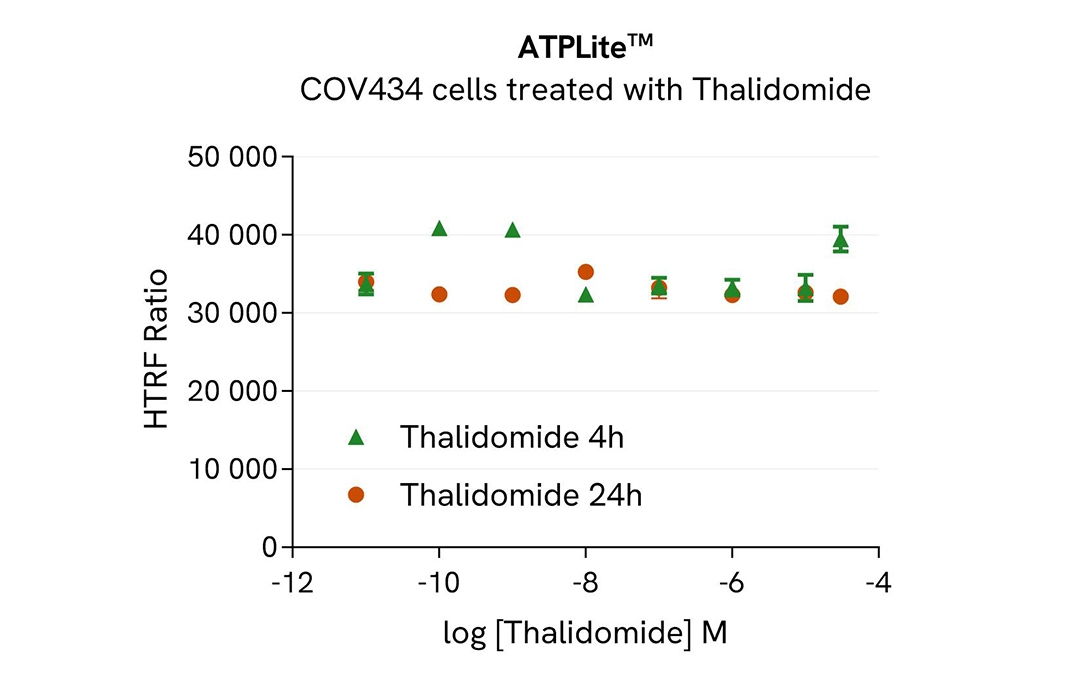
COV434 cells were seeded in 96-well culture plates at a density of 200,000 cells/well for 24 hours at 37°C, 5% CO2. Cells were treated with increasing concentrations of thalidomide for 4h or 24h.
After treatment, the cell culture medium was removed and 50 µL of supplemented Lysis Buffer#4 (1X) were dispensed into each well. Next, 16 µL of lysate were transferred into a 384-well low volume white microplate, and 4 µL of the HTRF Total SALL4 detection antibodies were added. An additional volume of lysate were also transferred into the microplate to check the alpha-tubulin level and cytotoxicity with the alpha-tubulin housekeeping Cellular Kit (64ATUBPET/G/H) and ATPlite Luminescence Assay System ( #6016736) respectively . HTRF signals were recorded after an overnight incubation.
The molecular glue Thalidomide induced a dose-dependent decrease in SALL4 protein levels with DC50* of 106 nM and 639 nM after 4h and 24h, respectively.
* DC50 corresponds to the concentration of the degrader at which 50% of the targeted protein is degraded.
Total SALL4 modulation using molecular glues is mediated by ubiquitin-proteasome system
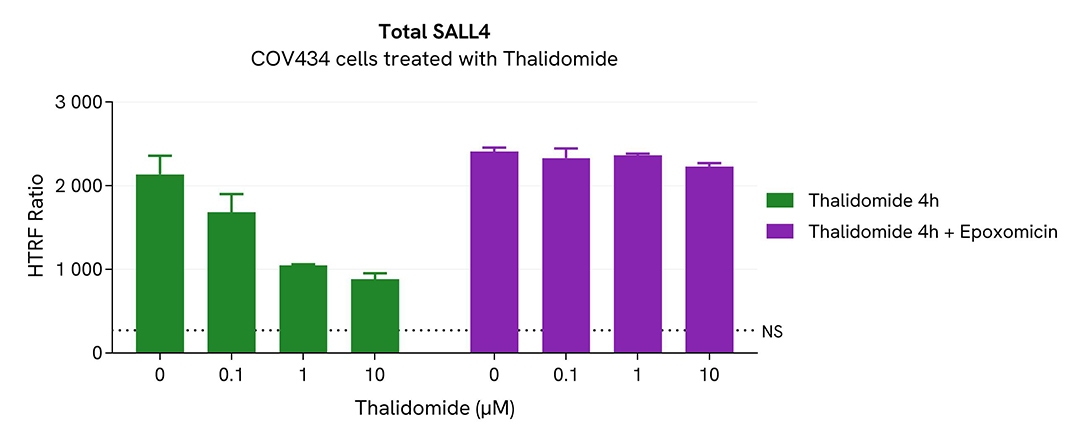
COV434 cells were seeded in 96-well culture plates at a density of 200,000 cells/well for 24 hours at 37°C, 5% CO2. Cells were then treated with increasing concentrations of the thalidomide for 4h. The proteasome inhibitor epoxomicin (1µM) was added or not 1h prior to the addition of Thalidomide.
After treatment, the cell culture medium was removed and 50 µL of supplemented Lysis Buffer#4 (1X) were dispensed into each well. Next, 16 µL of lysate were transferred into a 384-well low volume white microplate, and 4 µL of the HTRF Total SALL4 detection antibodies were added. An additional volume of lysate were also transferred into the microplate to check the alpha-tubulin level and cytotoxicity with the alpha-tubulin housekeeping Cellular Kit (64ATUBPET/G/H) and ATPlite Luminescence Assay System ( #6016736) respectively. HTRF signals were recorded after an overnight incubation.
Thalidomide-induced SALL4 degradation was prevented in the presence of epoxomicin, a proteasome activity inhibitor. This result unambiguously demonstrates that thalidomide-induced SALL4 degradation is mediated by the ubiquitin proteasome system.
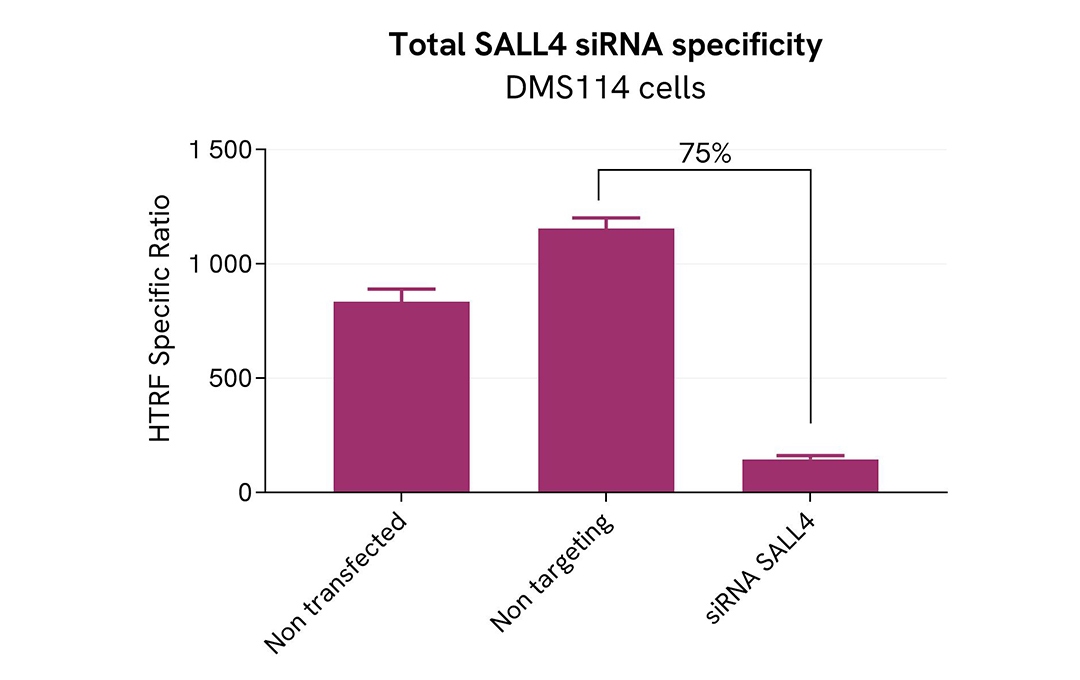
Validation of the selectivity of Total SALL4 assay using siRNA
DMS114 cells were plated in a 96-well plate (100,000 cells/well) and cultured for 24h. The cells were then transfected with 100nM siRNA specific to SALL4 as well as with a negative control siRNA. After 24 hours, the medium was replaced with fresh culture medium, and the cells were incubated for an additional 24 hours.
Post-treatment, the cells were lysed with 50 µL of supplemented lysis buffer #4 (4X) for 30 minutes at room temperature under gentle shaking. Subsequently, 16 µL of the lysates were transferred to a 384-well low-volume white microplate, and 4 µL of HTRF Total SALL4 detection antibodies were added. HTRF signals were recorded after overnight incubation.
Transfection with SALL4 specific siRNA resulted in 75% decrease in signal compared to cells transfected with the negative control siRNA. These results demonstrate the specificity of the HTRF total SALL4 assay.
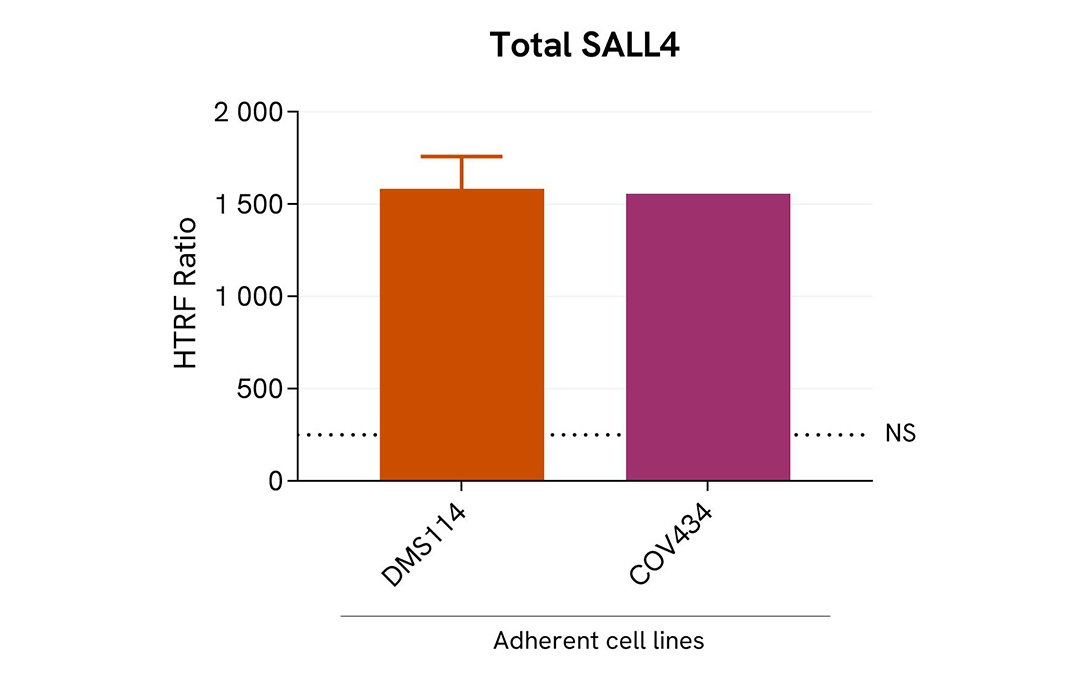
Assessment of Total SALL4 level in various cell lines
The adherent cell lines (COV434 & DMS114) were plated in 96-well culture plate and incubated for 24 hours at 37°C, 5% CO2. After medium removal, cells were lysed with 50 µL of supplemented lysis buffer #4 (1X) for 30 min at RT under gentle shaking.
The Total SALL4 expression level was assessed with the HTRF Total SALL4 kit. Briefly, 16 µL of cell lysate were transferred into a low volume white microplate, followed by 4 µL of premixed HTRF detection reagents. The HTRF signal was recorded after an overnight incubation at RT. The dotted line corresponds to the non-specific HTRF signal. Note that the cell density was optimized beforehand to ensure HTRF detection within the dynamic range of the kit (data are shown for 200,000 cells/well)
The HTRF Total SALL4 assay efficiently detected endogenous SALL4 in various cellular models expressing different levels of the protein.
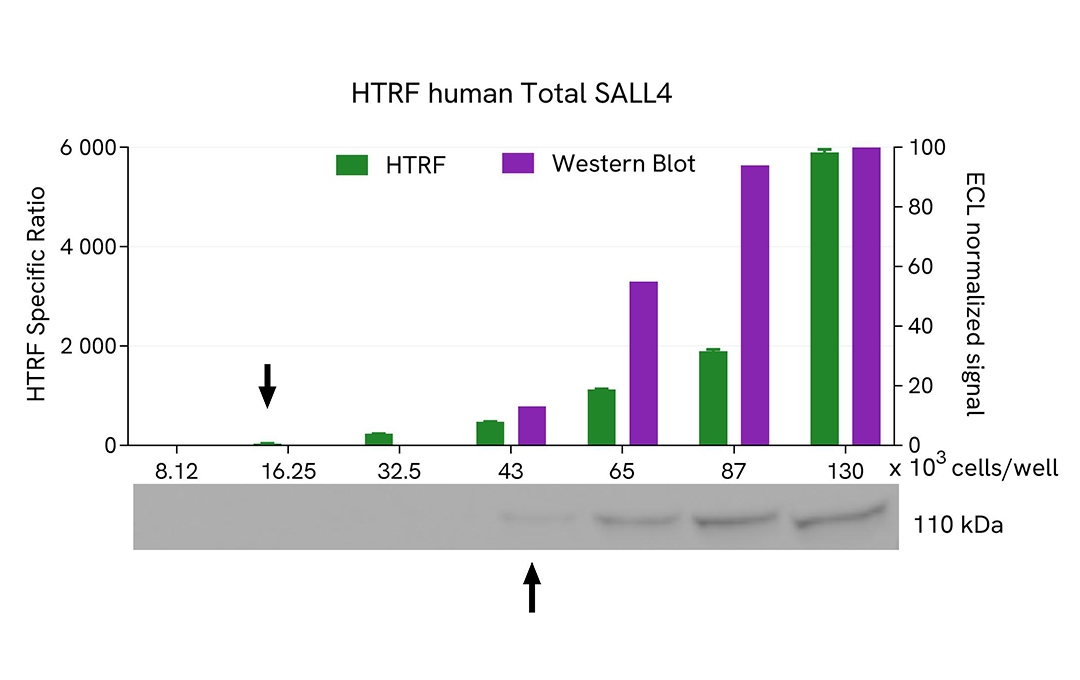
HTRF Total SALL4 assay compared to Western Blot
COV-434 cells were grown in a T175 flask in complete culture medium at 37°C - 5% CO2 until 80% confluence. After a 24h incubation, the cells were lysed with 3 mL of supplemented lysis buffer #4 (1X) for 30 minutes at RT under gentle shaking.
Serial dilutions of the cell lysate were performed using supplemented lysis buffer, and 16 µL of each dilution were transferred into a low volume white microplate before the addition of 4 µL of HTRF Total SALL4 detection reagents. Equal amounts of lysates were used for a side-by-side comparison between HTRF and Western Blot.
In these conditions, the HTRF Total SALL4 assay was 2 times more sensitive than the Western Blot technique.
Simplified pathway
SALL4 signaling pathway
SALL4 plays a key role in activating the Wnt/β-catenin signaling pathway by binding to the CTNNB1 promoter region and acting as a transcriptional activator, which enhances the expression of β-catenin. The β-catenin–TCF4 complex subsequently binds to TRIB3 and recruits SALL4. Additionally, SALL4 promotes the expression of Wnt3a and Bmi-1. Through these interactions, SALL4 activates the Wnt/β-catenin signaling pathway, leading to the upregulation of downstream target genes such as c-Myc, AXIN2, and CCND1. This cascade ultimately contributes to tumorigenesis and metastasis.

Loading...


How can we help you?
We are here to answer your questions.





































