In the rapidly evolving field of genomics, understanding gene expression at the translational level is becoming increasingly important. Ribosome profiling, or Ribo-Seq, has emerged as a powerful technique that provides a snapshot of ribosome positions on messenger RNA (mRNA) transcripts, revealing which genes are being actively translated. Interestingly, the library preparation methods used in small RNA sequencing (Small RNA-Seq) can be effectively adapted for Ribo-Seq, offering a streamlined approach to study translation comprehensively.

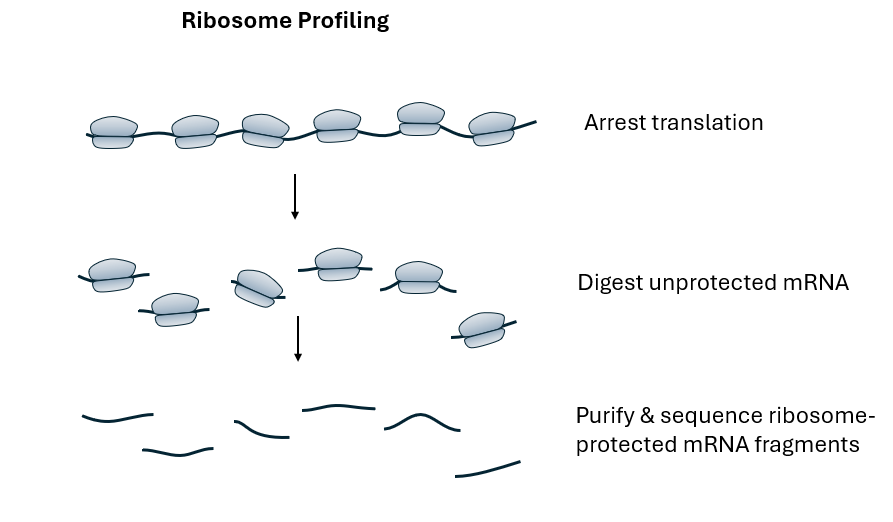
Figure 1: Diagram showing how mRNA transcripts are prepared for riboprofiling
Small RNA-Seq is a next-generation sequencing (NGS) technique designed to analyze small RNA molecules, typically less than 200 nucleotides in length, such as microRNAs (miRNAs), small interfering RNAs (siRNAs), and piwi-interacting RNAs (piRNAs). These small RNAs play critical roles in gene regulation. Ribosome profiling involves sequencing ribosome-protected mRNA fragments (RPFs) to provide insights into active translation. When ribosomes engage in protein synthesis, they protect a specific length of mRNA from nuclease digestion. By isolating and sequencing these protected fragments, researchers can determine which mRNAs are being actively translated and at what frequency.
The core similarity between Small RNA-Seq and Ribo-Seq lies in the size of the RNA fragments they analyze. In Ribo-Seq, the ribosome-protected fragments are approximately 28-30 nucleotides long, falling within the range of small RNAs. Therefore, the library preparation protocols developed for Small RNA-Seq are well-suited for preparing Ribo-Seq libraries. This adaptation involves key steps such as RNA fragment isolation, where mRNA-ribosome complexes are treated with nucleases to digest unprotected mRNA regions, leaving behind RPFs. This aligns with the extraction of small RNA fragments in Small RNA-Seq.
Adapter ligation is performed next, where specific adapters are ligated to the RNA fragments' 5' and 3' ends to facilitate reverse transcription and amplification. The adapters used in Small RNA-Seq are compatible with the size and ends of RPFs, making them suitable for Ribo-Seq library preparation. Reverse transcription and amplification then proceed as in Small RNA-Seq, converting the adapter-ligated RNA fragments into complementary DNA (cDNA) and amplifying them via polymerase chain reaction (PCR) to ensure sufficient material for sequencing. Size selection is also crucial, using methods like gel electrophoresis or bead-based purification to select for the desired fragment sizes, ensuring that only the ribosome-protected fragments are sequenced in Ribo-Seq.
Utilizing Small RNA-Seq library preparation in Ribo-Seq offers several benefits. It enhances efficiency and simplicity by streamlining the Ribo-Seq library preparation process, allowing laboratories familiar with Small RNA-Seq to adapt without significant changes to their workflows. It is cost-effective, leveraging existing reagents and protocols, which reduces the need for specialized kits. The high-throughput capability of gel-free Small RNA-Seq methods allows for the analysis of multiple samples simultaneously in Ribo-Seq studies. Technical consistency is maintained across different types of RNA sequencing, enhancing the reproducibility and comparability of results.
By adapting Small RNA-Seq library preparation for Ribo-Seq, researchers gain deeper insights into the translational landscape of the cell, with significant implications for translational research. This integration aids in gene expression analysis by identifying key regulatory mechanisms at the translational level. It has profound impacts on disease research, as aberrations in translation can lead to conditions such as cancer and neurodegenerative disorders; Ribo-Seq helps pinpoint these translational anomalies. In drug development, targeting translational control mechanisms offers new avenues for therapeutic interventions, and Ribo-Seq data can identify potential drug targets.
While the adaptation is beneficial, certain challenges and considerations must be acknowledged. Ribosome footprint complexity, such as ribosome stalling and stacking, can create footprints of varying lengths, requiring careful size selection during library preparation. Technical sensitivity is another concern, as small RNA fragments are susceptible to degradation, necessitating stringent RNA handling protocols. Additionally, the complexity of data analysis in Ribo-Seq requires specialized bioinformatics tools to accurately map ribosome positions and translate counts into meaningful biological insights.
In conclusion, the use of Small RNA-Seq library preparation methods in Ribo-Seq represents a strategic convergence of technologies that enhances our ability to study translation comprehensively. This approach not only maximizes resource utilization but also facilitates more accessible and scalable ribosome profiling experiments.
References:
- Ingolia, N. T., et al. (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science, 324(5924), 218-223.
- Guo, H., et al. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature, 466(7308), 835-840.Meyer, K. D., & Lecanda, A. (2020). Specialized ribosomes, RNA regulons, and RNA-binding proteins: New layers of complexity in gene regulation. Trends in Biochemical Sciences, 45(9), 757-768.treamlined approach to study translation comprehensively.