High-content screening technology has the potential to accelerate research by extracting huge amounts of information from images of cells, on a cell-by-cell basis.
However, many researchers analyze only a small number of the properties captured within their images which means they’re missing out on valuable information from their precious cells.
It’s not about making use of the multiplexing potential of a high-content screening approach, but about extracting information that’s already there. For example, virtually all screening assays use a nuclear counterstain such as Hoechst to facilitate segmentation of cells. This counterstain can provide a whole new perspective on your cells, at no additional cost.
As an example, we describe in an application note how you can use a nuclear stain to construct detailed multi-parametric phenotypic profiles of cells, and in turn, how these profiles can be used to distinguish different cell types within a co-culture system.
It’s quite usual to culture primary cells together with other cells – such as primary hepatocytes with fibroblasts, or primary neurons with astrocytes – to help keep them alive and healthy. Or you may set up a co-culture to investigate the interactions between cell types, e.g. cancer cells with tumor-derived fibroblasts or epithelial cells with lymphocytes. But how can you then analyze the different cell types separately?
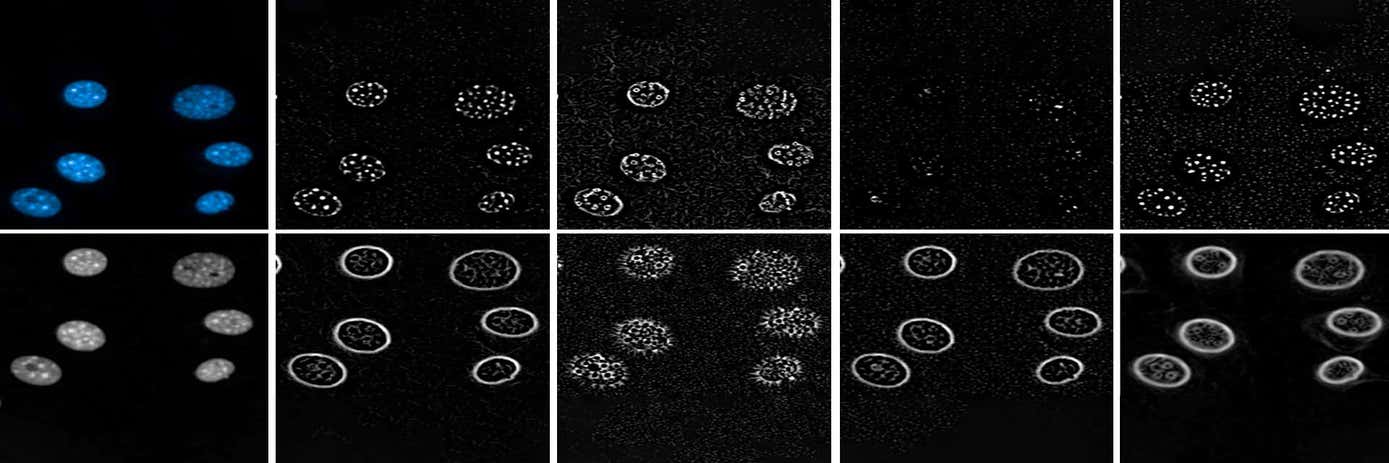
In one example, we co-culture human hepatocytes with mouse fibroblasts, and stain the nuclei with Hoechst. As well as measuring basic morphological properties (area, roundness, width, length) and intensities, we captured more sophisticated parameters, known as Spots, Edges, Ridges (SER texture), and Symmetry, Threshold compactness, Axial and Radial distribution (STAR morphology) of the nuclear stain. A total of 230 parameters were calculated for every nucleus.
Using a machine-learning approach, the best parameters to discriminate between the two cell types were selected, and then used to correctly classify cells either as HepG2 or NIH/3T3.
We found that the features necessary to distinguish the two cell types were all SER texture and STAR morphology properties, so to test the methodology further in a second example, we looked at whether these properties alone could separate even more cell types from one another. This time we took seven different cell lines and calculated the advanced SER and STAR properties. Principal component analysis showed clearly that these properties could be used to correctly classify the seven cell types.
But these are just examples. This type of phenotypic analysis can be applied to other cell types, and organelles other than the nucleus. Applying it to other fluorescent labels, or even cells labeled by a cell painting approach, opens up new possibilities for unbiased drug discovery and disease research.