Introduction to concentration and viability
Measuring viability and concentration of nucleated cells in clinica l research samples is inherently difficult due to the presence of red blood cells (RBCs) and cellular debris. In an attempt to remove the contaminating RBCs, the clinical research samples such as PBMCs, bone marrow and cord blood routinely undergo RBC lysis or Ficoll gradient separation. But even these methods are not always successful in removing all RBCs and directly lead to significant errors when determining cell concentrations. By using the Cellometer dual-fluorescence method, described below, we removed the RBC lysis and ficoll gradient steps. Consequently, we could measure the viability and concentration of primary clinical research samples without removing RBCs from the sample.
Cellometer dual-fluorescence counting method
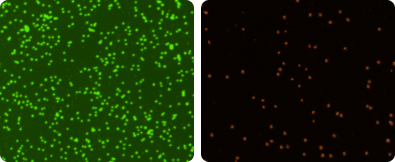
For viability determination, 20 µl of sample is mixed with 20 µl of Cellometer AO/PI Staining Solution. The acridine orange (AO) dye stains DNA in nucleated cells, generating green fluorescence. Propidium iodide (PI) stains DNA in cells with compromised cell membranes, generating red fluorescence. In cells stained with both AO and PI, the green fluorescence is absorbed by the red fluorescence via FRET (fluorescence resonance energy transfer). Dead nucleated cells fluoresce red (image far right). Live nucleated cells fluoresce green (image right). Live and dead cells are counted in the fluorescent images.
Red blood cells (RBCs), platelets, and debris do not fluoresce and are excluded from cell counts, so sample lysis is not required with the AO/PI viability method.

AO/PI stained cell images
Total nucleated cell count (TNC) viability for fresh clinical research samples
Cell Images of Fresh Clinical Research Samples
Because fresh clinical research samples contain a high percentage of platelets and red blood cells, incoming samples are sometimes not evaluated for TNC concentration. In some labs, samples are lysed for TNC analysis, but cell viability cannot be determined.
The Cellometer™ AO/PI dual-fluorescence method can be used to measure both TNC concentration and viability in fresh clinical research samples, such as bone marrow, cord blood, and peripheral blood without lysing.
AO/PI analysis procedure:
- Dilute bone marrow, cord blood or peripheral blood sample 1:10 with PBS.
- Combine 20 µl of diluted sample with 20 µl of AO/PI Staining Solution.
- Add 20 µl of stained sample to the Cellometer counting chamber and insert into a Cellometer automated cell counter such as the Cellometer Auto 2000 or Cellometer K2.
- The Cellometer system automatically captures and analyzes brightfield and fluorescent images.
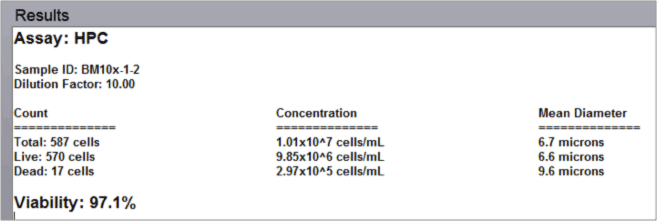
- The automated data report provides cell count, concentration, and mean diameter for both live and dead nucleated cells along with the % viability for the sample.
Fresh bone marrow aspirate analysis results
Brightfield image

All cells
Green fluorescent image
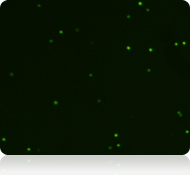
Live nucleated cells
Red fluorescent image

Dead nucleated cells
Data report
Fresh cord blood analysis results
Brightfield image

All cells
Green fluorescent image
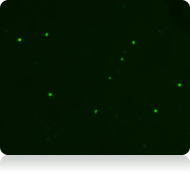
Live nucleated cells
Red fluorescent image

Dead nucleated cells
Data report

Fresh peripheral blood analysis results
Brightfield image
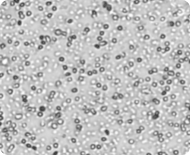
All cells
Green fluorescent image
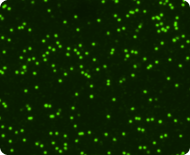
Live nucleated cells
Red fluorescent image

Dead nucleated cells
Data report
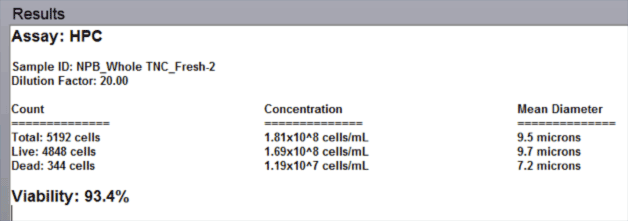
Reports are automatically displayed on the screen. A printed report with images can be generated directly from the Cellometer software
Analysis of mono-nuclear cells (MNC) and peripheral blood mononuclear cells (PBMC)
Following removal of RBCs and platelets via density gradient separation, samples are often counted manually using Trypan Blue staining to calculate viability. Because samples can contain varying amounts of RBC contamination following density gradient separation, using the AO/PI dual-fluorescence method for samples at this stage offers improved accuracy and reproducibility.
AO/PI analysis procedure:
- Combine 20 µl of sample with 20 µl of AO/PI Staining Solution.
- Add 20 µl of stained sample to the Cellometer counting chamber and insert into a Cellometer automated cell counter such as the Cellometer Auto 2000 or Cellometer K2.
- The Cellometer system automatically captures and analyzes brightfield and fluorescent images.
- The automated data report provides cell count, concentration, and mean diameter for both live and dead nucleated cells along with the % viability for the sample.
PBMC sample analysis results
Brightfield image

All cells
Green fluorescent image
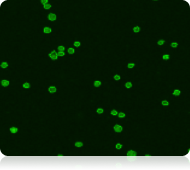
Live nucleated cells
Red fluorescent image
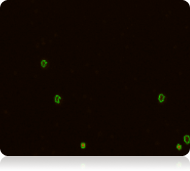
Dead nucleated cells
Data report
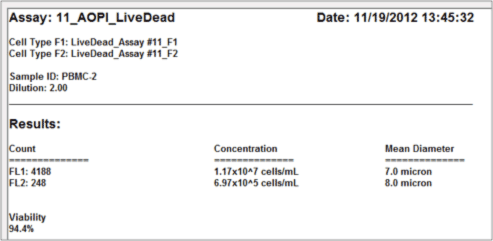
As demonstrated below, there can be RBC contamination in samples following density gradient separation.
Using the AO/PI method supports reduction of potential errors caused by counting of RBCs
Staining of nucleated cells in a PBMC sample
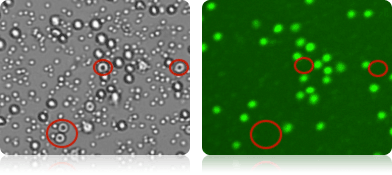
RBCs present in the PBMC sample are visible in the brightfield image acquired with a Cellometer Vision 10x instrument (far left). Because RBCs do not contain DNA, they are not stained by AO and are not counted as live nucleated cells in the green fluorescent image (near left).
Impact of RBC contamination
RBCs are not stained by AO/PI and are not identified by the Cellometer software

Red blood cells look like white blood cells under the microscope. They can often be included when manually counting TNCs.
PBMC brightfield image

Platelets, WBCs, and RBCs can be identified and counted in images taken with the Cellometer Vision instrument to measure the amount of RBC contamination in a sample. (The Cellometer Vision instrument has been superseded by the Cellometer Spectrum).
RBC contamination is present following density gradient separation
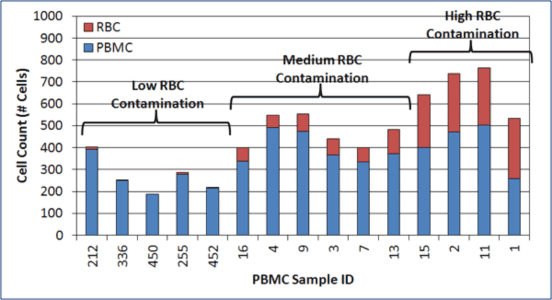
In 15 random PBMC samples evaluated, 4 samples showed a high degree of RBC contamination and 6 samples showed moderate RBC contamination.
RBC contamination is present in MNC and PBMC samples.
The amount of RBC contamination varies from one sample to another.
Manual counts overestimate PBMC counts & viability in samples with RBCs
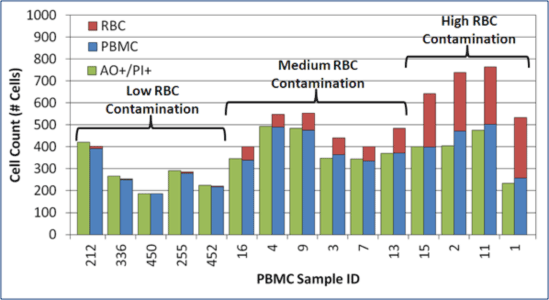
For samples containing high RBC contamination, manual counts over-estimated WBC concentration by ~25 to 50%.
AO/PI cell counts closely matched actual PBMC counts.
AO/PI provided a more reliable PBMC concentration and viability in samples with RBC contamination.
Analysis of isolated cell populations from purified clinical research samples
Following immune-magnetic separation, samples display a high degree of purity and can be counted using a wide range of counting methods, including both Trypan Blue and AO/PI. Whether isolated from bone marrow, cord blood, or peripheral blood, isolated cell populations are typically clean, with no RBC or platelet contamination. Several examples of specific cell populations isolated from different types of samples are shown below.
CD56+ NK Cells from NPB

CD34+ Stem cells from CB
CD19+ B cells from NPB
AO/PI correlation to manual counting for clinical research samples
As part of a collaboration with AllCells , a provider of primary cells, two popular brightfield counting methods were compared to Cellometer AO/PI dual-fluorescence counting. Results correlated well for both the Methylene Blue counting method (for all sample types) and the Trypan Blue counting method (for isolated cell populations).
Such correlation to manual counting supports labs with implementing the change to the Cellometer AO/PI counting method with minimal documentation and testing.
AO/PI and methylene blue TNC counts correlated well for fresh samples
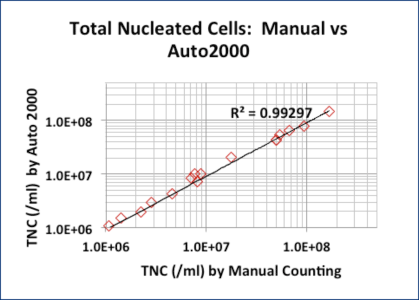
Sixteen samples (peripheral blood, cord blood, and bone marrow aspirates) were lysed / stained with 3% acetic acid / methylene blue and counted manually.
A second aliquot of each sample was stained with AO/PI without lysing and counted with the Cellometer Auto 2000.
Though the 3% acetic acid / methylene blue method provides accurate TNC counts, it does not measure cell viability.
The AO/PI method does not require cell lysis.
Trypan blue and AO/PI counts correlated well for purified samples
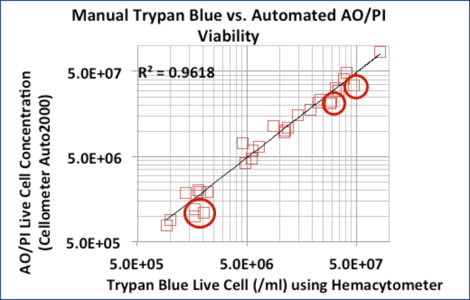
Correlation between Trypan Blue and AO/PI for 24 purified samples (CD14+, CD19+, and CD34+ cells)
6 MNC samples did not correlate as well. Trypan Blue counts were higher due to RBC contamination. (red circles).
Trypan Blue or AO/PI can be used to measure concentration and viability in isolated cell populations, such as CD34+, CD56+, and CD19+ cells.
For research use only. Not for use in diagnostic procedures.




























